Abstract
Available ecological information, an extensive distributional range, conflicting osteological data, and a proposed early Miocene origin provide the impetus for the present study which investigates genetic structuring, biogeographic, and phylogenetic relationships within the Aplocheilichthys spilauchen lineage. Through the analysis of the mitochondrial gene COI, species delimitation methods (ABGD, GB, GMYC, bPTP) were applied, recognizing 6–7 OTUs with absolute pairwise genetic distances ranging between 8 and 22%. The onset of diversification is estimated to be within the middle Miocene and both dispersal and vicariance-shaped A. spilauchen diversity and distribution, as suggested by time-calibrated and ancestral range reconstruction (S-DIVA) analyses. We report for the first time, a pattern of diversification within a lineage of brackish water fish that is concordant with the historical distribution of coastal mangroves forests, shaped by a series of historical events that likely affected forest cover since the middle Miocene (e.g. major climate shifts and sea-level fluctuations, onset of the modern Congo River outlet, increased volcanism in the Cameroon Volcanic Line).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mangrove forests are among the most dynamic habitats in tropical and subtropical intertidal zones, where they are limited to low-lying coastal areas with constant input of freshwater (Kathiresan & Bingham, 2001; Duke, 2017). Along the African Atlantic coast, the latitudinal extent of mangroves is constrained by two cold sea currents, the Benguela Current in the south, flowing northwards from South Africa to Angola, and the Canary Current flowing southwards from Morocco to Senegal (Saenger & Bellan, 1995) (Fig. 1). The presence and extent of mangrove stands in Africa are directly related to humidity, rainfall, and coastal relief and are known to have undergone extensive expansions and contractions in both range and abundance over time (Saenger & Bellan, 1995). Indeed, the study of mangrove cover is a primary source of information for modelling impacts of climate change (Alongi, 2015), as well as for palaeoclimatic reconstructions based on historical data from mangrove pollens (Bonefille et al., 1982; Leroy & Dupont, 1994; Scourse et al., 2005; Durogbo et al., 2010; Adeonipekun et al., 2016; Vallé et al., 2017).
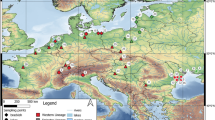
Map of West and West-Central Africa with mangrove distribution (light green) and Aplocheilichthys spilauchen sampling sites (black dots) indicated. AC Angola Current, AR Atlantic Rise (red), BC Benguela Current, CC Canary Current, CVL Cameroon Volcanic Line (yellow bar), FP Freshwater plume (blue), GC Guinea Current, GGC Gulf of Guinea Current, SEC South Equatorial Current. Arrows indicate currents direction
The Atlantic coast of Africa has an extremely narrow continental shelf, resulting in mangrove expansion during high sea level but contraction during low sea level (Saenger & Bellan, 1995; Scourse et al., 2005). In a high sea-level scenario, mangroves expand over new floodplains along the lower sections of rivers newly under tidal influence and salinity. In the opposite low sea-level scenario, fewer surfaces are exposed and mangroves recede to the newly reduced tidal zone, with the original stands replaced by other vegetation types. This is in contrast to regions with wide continental shelves, such as in Southeast Asia where significant mangrove expansion is related to low sea level, as shallow shelf areas previously covered by saltwater are exposed to tidal influence and freshwater inputs (Luther & Greenberg, 2009). A similar pattern is seen along the Brazilian coast, also with a wide continental shelf, where during periods of low sea level, many brackish water palaeochannels form and connect adjacent river drainages providing ideal conditions for mangrove expansion (Thomaz et al., 2015; Thomaz & Knowles, 2018). Similar to the described mangrove dynamics, the distribution of brackish water fishes is also highly dependent on freshwater inputs from effluent river systems and sea-level variations (Saenger & Bellan, 1995; Kathiresan & Bingham, 2001; Duke, 2017). Such alternating expansion and contraction dynamics can reveal interesting historical information when analysed in phylogenetic and temporal contexts, and this is particularly the case for taxa inhabiting regions known for major shifts in river outflows, climatic instability, sea level changes, and tectonic activity such as along the African Atlantic coast (Giresse, 2005; Goudie, 2005; Runge, 2007; Flugel et al., 2015).
Among the procatopodids, a cyprinodontoid group comprising mainly freshwater fishes endemic to Africa, the banded lampeye, Aplocheilichthys spilauchen (Duméril, 1861), is one of the few brackish water species and is a dominant species in mangrove habitats along the tropical and subtropical zones of the Atlantic coast (Fig. 1). Similar to other procatopodids and many other cyprinodontoids, A. spilauchen is known for a well-developed swimming capability (vagility) when compared to members of the Aplocheiloidei that in Africa are represented by the Nothobranchiidae (Huber, 1999). The latter is found mostly in shallow freshwater habitats, and some taxa even exhibit a seasonal life cycle (e.g. Nothobranchius), thus, presenting a completely different response to climatic and biogeographic processes. A river or an increase in the input of freshwater into an estuarine region likely represents range expansion opportunities for cyprinodontoid taxa, whereas for aplocheiloids, they more likely act as barriers (Bartáková et al., 2015).
Although A. spilauchen has also occasionally been collected from inland freshwater habitats, it primarily has a coastal distributional range, which closely mirrors the area covered by Atlantic coast mangrove forests, from the Senegal River to the Kwanza River in Angola (Wildekamp, 1995; Saenger & Bellan, 1995; Feka & Morrison, 2017) (Fig. 1). Aplocheilichthys spilauchen is currently recognized as the sole member of Aplocheilichthys and was the first African lampeye to be described (as Poecilia spilauchen), based on specimens from the Ogowe River in Gabon by Duméril (1861). It is readily distinguished from all other procatopodids by the presence of a combination of vertical grey banding along the flanks and dorsal, anal, and caudal fin in males and attainment of large body size (Fig. 2). Adults routinely reach up to 7.0 cm standard length while other procatopodids rarely exceed 4.0 cm standard length (Wildekamp, 1995). Likely, due to this distinctive pigmentation patterning and its presence in mangroves and other brackish water habitats, it has been assumed that A. spilauchen is a widely distributed species. Two other species, A. typus Bleeker, 1863 probably from Ghanaian coastal regions (Wildekamp, 1995), and A. bensonii (Peters, 1864) from Liberia were described but promptly synonymized with A. spilauchen by Günther (1866). A third species, A. tschiloangensis Ahl, 1928, described from the Tschiloango River in Cabinda, an Angolan territory north to the Congo River outlet, is now also considered a synonym of A. spilauchen (Huber, 1982; Wildekamp et al., 1986).
Males of A. spilauchen from localities along the Atlantic coast of Africa: a UFRJ 4150, 37.6 mm SL, Sangrougrou River, Senegal; b AMNH 275472, 51.0 mm SL, Farmoriah River, Guinea; c AMNH 59382, 52.8 mm SL, Freetown, Sierra Leone; d UFRJ 11487, 44.8 mm SL, Assimie, Ivory Coast; e MRAC 73–08-P-135–138, 41.0 mm SL, Ebrie Lagoon, Ivory Coast; f AMNH 226603, 46.4 mm SL, coastal Benin; g UFRJ 11484, 42.3 mm SL, coastal Nigeria (aquarium import); h MRAC 143378–387, 64.7 mm SL, Bioko Island, Equatorial Guinea; i MRAC 73–39-P-2168–179, 41.4 mm SL, mouth of Mirupururu River, Bioko Island, Equatorial Guinea; j AMNH 258331, 45.5 mm SL, mouth of Kouilou River, Republic of Congo; k AMNH 238526, 31.0 mm SL, Boma estuary, lower Congo River, Congo DRC; l SAIAB 84605, 40.0 mm SL, Dondo, Kwanza River, Angola
Investigating seasonal variations in mangrove fish communities in Nigeria, Wright (1986) found that A. spilauchen was the dominant species, both in numbers and biomass and that its reproductive cycle was dependent upon freshwater inputs. Okyere (2012), in a study evaluating the use of A. spilauchen for mosquito larvae control along the Ghanaian coast, found that the species exhibited a low resilience to salinity increase, tolerating only a maximum of 4‰ salinity (Okyere, 2012). Both ecological studies suggest a dependence on freshwater in shaping the niche requirements and distribution of the species. Osteological information available for A. spilauchen reveals some inconsistencies regarding the configuration of the hypural plate supporting the caudal fin, usually an invariant feature among procatopodid species. The plate was considered as completely fused (Parenti, 1981), separated (Ghedotti, 2000), or bearing a small gap close to the compound caudal centrum (Costa, 2012).
Recently, in the first molecular dated analysis focused on the Procatopodidae, A. spilauchen was resolved as the sole member of a lineage sister to all other procatopodids, except Plataplochilus Ahl, 1928 (Bragança & Costa, 2019). Through the inclusion of two fossil calibrations in the sister families Valenciidae and Aphaniidae, it was estimated that A. spilauchen split from the remaining procatopodids in the early Miocene (around 23 mya). Considering the diversification patterns seen in all other procatopodid lineages, which intensified during the middle/late Miocene and Pliocene, the presumed lack of diversification within the A. spilauchen lineage is worthy of investigation.
Available ecological information (Wright, 1986; Okyere, 2012), conflicting osteological data (Parenti, 1981; Ghedotti, 1998, 2000; Costa, 2012) and a proposed early Miocene origin for the A. spilauchen lineage (Bragança & Costa, 2019) provide the impetus for the present study which aims to (1) investigate cryptic diversity within the A. spilauchen lineage through the application of both distance and coalescent species delimitation methods, (2) examine connectivity or genetic structuring between haplotypes, (3) estimate the divergence times for identified lineages, (4) perform ancestral area reconstructions, and (5) interpret the resulting temporal and biogeographic patterns in view of the current knowledge of the main historical events that have affected the African landscape and influenced Atlantic mangroves and coastal habitats.
Material and methods
Specimen preservation and fixation
Samples were collected using a variety of fishing techniques, and specimens were euthanized with clove oil or MS-222 anaesthetic solutions, in accordance with recommended guidelines for the use of fishes in research (Bennett et al., 2016). A small piece of muscle tissue or fin was taken from the right side of each specimen, or the entire fish was preserved in 95% ethanol in the field. The samples were stored at low temperatures at each of the following institutions: (1) NRF-South African Institute for Aquatic Biodiversity (NRF-SAIAB), Makhanda (Grahamstown), South Africa; (2) Ichthyological Collection of the Biology Institute of the Federal University of Rio de Janeiro (UFRJ), Rio de Janeiro, Brazil; (3) Royal Museum for Central Africa (RMCA), Tervuren, Belgium; (4) American Museum of Natural History (AMNH), New York, USA; (5) National Museum of Natural History (USNM), Washington, USA and (6) Oregon State University (OS), Corvallis, USA.
Taxon sampling
Samples of newly generated and published sequences of the mitochondrial gene COI (Cytochrome c oxidase subunit 1) with a total of 679 bp from eight populations of A. spilauchen from across its geographical range were included in the analysis (Fig. 1). The present dataset includes samples from the (1) Kwanza and Lucala rivers in Angola (n = 7); (2) lower Congo River in the Democratic Republic of Congo (n = 2); (3) Kouilou River in the Republic of Congo (n = 2); (4) Ngounie River (Ogowe River drainage) and Komo river in Gabon (n = 3); (5) Ntem River in Equatorial Guinea (n = 3); (6) Aquarium import specimens from coastal Nigeria (n = 2); (7) Kakum River in Ghana (n = 3) and (8) Forecariah River at Conakry, Guinea (n = 2). Following Bragança & Costa (2019), three species were selected as outgroups: Plataplochilus miltotaenia Lambert, 1963 and P. pulcher Lambert, 1967, representing the first diverging lineage of the Procatopodidae and ‘Poropanchax’ normani (Ahl, 1928), representing the remaining procatopodid lineages sister to Aplocheilichthys. The generic name of ‘Poropanchax’ normani is between quotation mark to indicate that the species is not a member of Poropanchax sensu stricto as revealed in Bragança & Costa (2019), and awaits future taxonomic and nomenclatural resolution. A list of all included specimens and their respective catalogue numbers, localities, and GenBank Accession numbers are provided in Table 1. Other specimens, not included in the molecular analyses but photographed to illustrate pigmentation and body shape variation in different populations along the African coast, are shown in Fig. 2. The catalogue number and locality information of the specimens in Fig. 2 are listed in Online Resource 1.
DNA extraction and sequencing
DNA was extracted from preserved tissues using the salting out method (Sunnucks et al., 1996), or the DNeasy Blood & Tissue Kit (Qiagen, Hilden) following the manufacturer’s standard protocol for animal tissue isolation. A fragment of the mitochondrial gene COI (Cytochrome c oxidase subunit 1) was amplified with universal primers LCO1490 and HCO 2198 and published protocols (Folmer et al., 1994). PCR products were purified with Exosap (Applied Biosystems), cycle sequenced using BigDye Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and sequenced at the NRF-SAIAB using an ABI 3730xl DNA Analyzer (Applied Biosystems), or by Macrogen, South Korea.
Phylogenetic analyses, COI codon partitioning and evolution models
Phylogenetic analyses included newly generated and published sequences of the mitochondrial gene COI from A. spilauchen specimens from along the western coastal plains and mangroves of the Atlantic coast (see Table 1). Our COI alignment did not have frameshifts, indels or premature stop-codons, which are indicative of pseudogenes. The best models of evolution for each codon position for the maximum likelihood (ML) and Bayesian inference (BI) analyses were determined in PartitionFinder2 (Lanfear et al., 2016). The partitioned ML analysis was completed with GARLI (Zwickl, 2006), and bootstrap support was assessed from a majority rule consensus tree generated in Mesquite (Maddison & Maddison, 2019) from 1000 trees. The partitioned BI analysis was performed with MrBayes version 3.2 (Ronquist et al., 2012), and posterior probabilities were assessed with 5 million generations, sampling trees every 1000 generations. The first 25% of trees were discarded as burn-in. Both the ML and BI analyses were performed on the CIPRES Science Portal (Miller et al., 2010). The models of evolution implemented for each codon position in the ML and BI analyses were TRNEF + I + G, F81 + I, and TIM + G and SYM + G, F81 + I, and GTR + G, respectively.
For the species delimitation methods (described below), an ultrametric tree including only unique haplotypes (n = 15) as required by the GMYC method was performed in Beast 1.8.2 (Drummond et al., 2012), and the parameters were defined as follows: an uncorrelated relaxed clock model with a lognormal distribution (Drummond, et al., 2006), with 10 million generations with a sampling frequency of 1000. The value of parameters of the analyses, convergence of the MCMC chains, sample size and the stationary phase of chains were evaluated using Tracer 1.6 (Rambaut et al., 2014). A Birth and Death Incomplete Sampling speciation process for the tree prior (Stadler, 2009), indicated for datasets with incomplete sampling, was used, and the analysis included only ‘Poropanchax’ normani as outgroup. For this analysis, the evolution model HKY + G was inferred in JModeltest (Darriba et al., 2012) for the entire COI gene. Saturation levels were checked in Dambe5 (Xia, 2013), according to the algorithm proposed by Xia et al. (2003).
Species delimitation
Four species delimitation methods were applied to investigate the diversity within A. spilauchen. The traditional barcoding genetic distance method (GD) (Herbert et al., 2003) and the Automatic Barcode Gap Discovery (ABGD) (Puillandre et al., 2012), both relying on genetic distances between haplotypes, and the General Mixed Yule Coalescent (GMYC) (Fujisawa & Barraclough, 2013) and Bayesian implementation of the Poisson Tree Processes (bPTP) (Zhang et al., 2013), both coalescence approaches.
For the GD method, we used the Kimura-2-parameters model (K2P) (Kimura, 1980) to estimate the pairwise genetic distances between the different A. spilauchen populations in MEGA 7 software (Kumar et al., 2016). Haplotypes with a genetic divergence higher than 3% were considered as belonging to distinct operational taxonomic units (OTUs) following the expected pattern for genetic divergence between fish species (Ward, 2009). ABGD also relies on genetic distances to identify the threshold between interspecific (speciation) and intraspecific (populational) processes within the dataset (Puillandre et al., 2012). The main difference between GD and ABGD is that the ABGD allows a more refined search; once the estimated genetic divergence between groups (putative species) is calculated, it is recursively applied to the previously delimited groups until no more putative OTUs are recognized. The ABGD result is presented relative to a spectrum of P values (prior intraspecific values) in which a 0.001 value assumes a minimum intraspecific variability, and a 0.1 value assumes a maximum intraspecific variability. The different results relative to each significant P value intervals are presented, and to identify which P value interval best describes the diversity within the dataset, a congruence with other methods is expected and/or a support from traditional morphological alpha taxonomy. The ABGD analysis was performed in the ABGD server website (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html) following the default parameters.
The coalescence species delimitation method, GMYC (Fujisawa & Barraclough, 2013), requires an ultrametric tree to distinguish between intraspecific (coalescent) and interspecific (speciation) threshold patterns, because it relies on branch length differences to define OTUs. The bPTP coalescent method (Zhang et al., 2013), in contrast to GMYC, relies on the number of nucleotide substitutions between haplotypes to resolve intraspecific and interspecific patterns. When performing bPTP, two results are provided, the BI and the ML solutions, and in each, the recognized OTUs are depicted graphically in a tree. The same reduced dataset tree as for GMYC was used when performing bPTP. The GMYC analysis was performed on the Exelixis Lab's server https://species.h-its.org/gmyc/ following default parameters, and the bPTP was performed on the Exelixis Lab's server https://species.h-its.org/ptp/ following default parameters except for a 20% burn-in.
Haplotype network
The haplotype network was inferred, using the Minimum Spanning Network method (Kruskal, 1956) in PopART 1.7 (Leigh & Bryant, 2015), including only A. spilauchen haplotypes. The following eight geographic areas/drainages were delimited: (A = Gabon, B = Angola, C = Guinea, D = Kouilou, E = Lower Congo, F = Nigeria, G = Ghana, H = Equatorial Guinea). These areas were delimited based on the presence or absence of connectivity between haplotypes as suggested by the genetic divergence and species delimitation results. Therefore, the area ‘Angola’ includes all haplotypes from the Kwanza and Lucala rivers, the latter being one of the main tributaries of the lower Kwanza River. The area ‘Gabon’ includes all haplotypes from the Ngounie River, a tributary of the lower Ogowe River, and from the Komo River.
Time-calibrated analysis
The time-calibrated analysis was constructed in BEAST v.1.8.2 (Drummond et al., 2012), including all sequences available, and an uncorrelated relaxed clock model with a lognormal distribution (Drummond et al., 2006). Bayesian Inference was performed with 50 million generations with a sampling frequency of 1000. The values of parameters of the analysis, convergence of the MCMC chains, sample size, and the stationary phase of chains were evaluated using Tracer 1.6 (Rambaut et al., 2014). The same Birth and Death Incomplete Sampling speciation process was selected for the tree prior (Stadler, 2009). Since no fossil procatopodids are currently known three relevant calibration points based on the age estimates from Bragança & Costa (2019) were selected. These secondary calibrations correspond to (1) the Procatopodidae basal node (prior setting: normal distribution, mean = 30.1, standard deviation = 1.3); (2) the node representing the split between the A. spilauchen clade and ‘Poropanchax’ normani (prior setting: normal distribution, mean = 23.1, standard deviation = 1.5); and (3) the node representing Plataplochilus diversification (prior setting: normal distribution, mean = 3.2, standard deviation = 0.9). A normal distribution prior was selected for the secondary calibration points as recommended by Ho (2007) and Ho & Phillips (2009). At the end of the analysis, a burn-in of 25% of the retained trees was performed in TreeAnnotator.
Ancestral area reconstruction
The ancestral area reconstruction method, S-DIVA (Statistical Dispersal-Vicariance Analysis) (Yu et al., 2010), was performed in RASP 3.2 (Yu et al., 2015) to reconstruct the expected ancestral areas during A. spilauchen diversification. S-DIVA is an event-based method that incorporates statistical uncertainty into both phylogenetic and ancestral state reconstructions and assumes vicariance as the most probable event to explain biogeographical evolution within a group, attributing higher costs for dispersal and extinction events. For the S-DIVA analysis, both extinctions and reconstructions were considered, and at each node, the frequency of alternative ancestral area reconstructions generated for each tree in the dataset is shown. The analysis was performed on the resulting trees from the time-calibrated analysis from Beast 1.8.2 (Drummond et al., 2012), and the condensed tree was defined as the summary tree from that same analysis. Prior to the analysis, the outgroups were removed in RASP 3.2, and a post-burning of 12,500 trees was carried out. The maximum number of areas included in ancestral distributions was set to 3, but based on current knowledge from ecology, distribution, and genetic divergence between populations, only ancestral distributions between contiguous areas were allowed.
Results
Phylogenetic analysis (Online Resource 2)
ML and the BI analyses recovered the same topology with most nodes receiving posterior probability values > 95 and bootstrap values > 80, except for nodes representing the most recent divergences (Online Resource 2). An early divergence of Plataplochilus species and a sister group relationship between the A. spilauchen lineage and ‘Poropanchax’ normani were recovered with strong support. Within Aplocheilichthys, a sister group relationship between haplotypes from Ghana and Guinea was supported with a high posterior probability but a comparatively lower bootstrap value (Online Resource 2). Haplotypes from Gabon were resolved as sister to all remaining haplotypes with high support. Another node, also strongly supported, represents the split between Nigerian + Equatorial Guinean haplotypes from those from Kouilou, lower Congo, and Angola. A sister group relationship between Nigerian and Equatorial Guinean haplotypes was supported by maximum values in both analyses. The node-grouping haplotypes from Kouilou, lower Congo and Angola were supported with high posterior probability but a comparatively lower bootstrap value. However, within that clade, all nodes received lower support values, except the node representing the split between Kouilou haplotypes and those from lower Congo and Angola, which received strong support.
Species delimitation (Fig. 3)
Both genetic distance methods provided similar results, but as expected, with a decrease in intraspecific variability value (P value) in ABGD more OTUs were delineated. The ABDG result with a P value between 0.0129 and 0.0599 recovered the same six OTUs as delimited by the GD method: (1) Ghana, (2) Guinea, (3) Gabon, (4) Nigeria, (5) Equatorial Guinea, (6) Kouilou + lower Congo + Angola, whereas one more OTU was identified between P values of 0.0046 and 0.0077, with the Kouilou lineage considered as distinct from a lower Congo + Angola OTU. The absolute pairwise genetic distances for the OTUs identified based on the GD method range from 8 to 22%, which are considerably higher than the heuristic standard threshold of 2–3% commonly applied to recognize species limits (Table 2). The highest genetic distance (22%) was between specimens from Guinea and Equatorial Guinea while the lowest (2%) was recorded for specimens from Kouilou, lower Congo and Angola.
The Bayesian ultrametric tree (Fig. 3) had the same topology as the phylogenetic analyses (Online Resource 2), with most nodes supporting putative OTUs with maximum posterior probability values. There was concordance of the six OTUs defined by ABGD (P value between 0.0129 and 0.0599), GD, GMYC and the bPTP ML solution result, while bPTP BI solution returned the same seven OTUs as ABGD when assuming the lowest P values. We considered the six OTUs defined by the traditional barcoding, the ABGD including P values between 0.0129 and 0.0599, the GMYC and the bPTP ML solution as putative species but defer making any taxonomic changes pending a detailed osteological and morphometric study (see Discussion).
Haplotype network (Fig. 4b)
A minimum spanning network portraying genealogical relationships among haplotypes revealed considerable divergence between geographically separated populations within A. spilauchen. In most cases, the number of nucleotide substitutions separating populations was extremely high ranging from a minimum of 43 up to 133. The only exception to this being among haplotypes from the neighbouring Kouilou, lower Congo, and Angolan populations which form a cluster with a maximum of 12 nucleotide changes separating Kouilou haplotypes from the Angolan and lower Congo haplotypes.
Time-calibrated analysis (Fig. 5)
Time-calibrated phylogeny of the Procatopodidae and A. spilauchen obtained from the Bayesian dating analysis in BEASTv.1.8. Bars represent maximum and minimum date estimates for each node (values in brackets), and the numbers are nodes divergence mean ages. The colours in the time bar are a reference to the proposed time extension of main palaeogeographic and palaeoclimatic events during A. spilauchen evolution: the green bar represents the Miocene Climatic Optimum; the red bar represents the Miocene Climatic Transition; the orange bar represents the late Miocene aridification; the brown bar represents the Pliocene–Pleistocene climatic instability; the blue bar represents the onset of the modern Congo River outlet and the black bar represents an increase in the volcanic activity of the CVL
As expected, given a single marker dataset and uncertainties inherent in the use of secondary calibrations, most nodes on our time tree have 95% HPD values spanning large time intervals, and there is no question that considerable uncertainty persists regarding the precision of the resultant dating scheme. Nonetheless, despite these caveats, our scheme does provide a broad estimate of comparative divergence times among geographically disparate populations of A. spilauchen (Fig. 5). Based on the current analysis, the onset of diversification within the A. spilauchen lineage began during the Middle Miocene (14.8 mya, 95% HPD 8.6–20.7 mya), represented by the split of a west African lineage comprising specimens from Ghana and Guinea. These two lineages subsequently diverged during the late Pliocene (3.5 mya, 95% HPD 0.6–8.7 mya). A split between specimens from Gabon and all the remaining populations occurred during the late Miocene (9.5 mya, 95% HPD 4.8–14.9 mya). This was followed by another in the transition between the late Miocene and the Pliocene (5.6 mya, 95% HPD 2.3–9.5 mya), in which two clades diverged, one including specimens from Nigeria and Equatorial Guinea and the other those from the Kouilou, lower Congo and Angola. Later, the lineage including Nigerian and Equatorial Guinean specimens diverged at the onset of the Pleistocene (2.5 mya, 95% HPD 0.8–5.0 mya). The remaining lineage diversification time estimates are extremely recent ranging from around 1 mya to the near present.
Ancestral area reconstruction (Fig. 6)
Biogeographic ancestral area reconstruction inferred the current distribution pattern of A. spilauchen lineages to have likely resulted from repeated dispersal and vicariance events (Fig. 6). S-DIVA postulates 6 dispersal, 6 vicariance, and 1 extinction event. The root node, corresponding to the onset of diversification of the A. spilauchen lineage, estimated the areas FGH (Ghana, Nigeria and Equatorial Guinea) and CFG (Guinea, Ghana, and Nigeria), as equally most probable ancestral areas (49% probability). From this root node, a vicariance event is suggested between areas AFH (Gabon, Nigeria, and Equatorial Guinea) and CG (Guinea and Ghana). At the node corresponding to the split between Guinean and Ghanaian haplotypes, the ancestral area CG was recovered (96% probability), and a vicariance event was suggested as the cause of the split between the two areas. The ancestral area AFH was identified for the node in which the Gabon haplotypes split from the remaining haplotypes (95% probability), and it is possible to identify a duplication event (within area speciation) in area A (Gabon) followed by a vicariance event in which one lineage is restricted to Gabon and the other present in area AFH. The area AFH was considered as the ancestral area for the node in which Nigeria and Equatorial Guinea haplotypes split from the Kouilou, lower Congo and Angolan haplotypes (93% probability). Vicariance was recovered as the explanation for the split between areas FH (Nigeria and Equatorial Guinea) and ADE (Gabon, Kouilou, and lower Congo). The ancestral area FH was estimated for the node in which Nigeria and Equatorial Guinea haplotypes split (96% probability), caused by a vicariance event. For the node which corresponds to the split between Kouilou haplotypes and Angola + lower Congo haplotypes, the ancestral area ADE (Gabon, Kouilou, and lower Congo) (93% probability) was estimated, followed by an extinction event in area A (Gabon), a dispersion to area B (Angola) and finally a vicariance event between areas BE (Angola and lower Congo) and D (Kouilou). For the split between Angola and lower Congo haplotypes, the ancestral area BE was delimited (100% probability) and considered to be the result of a vicariance event.
Discussion
Prior to the current study, A. spilauchen had been considered to be widely distributed with a range extending along much of coastal west and west-central Africa. This was based on the assumption that a higher salinity tolerance, relative to that of other procatopodids, would have allowed the species to maintain population connectivity across this extensive geographical range. In addition, an apparent lack of variability in pigmentation patterning between individuals from geographically disparate populations supported a widespread species hypothesis, which in turn resulted in no further investigation of morphological differences between them. Unfortunately, the ongoing COVID-19 pandemic has prevented us from undertaking a morphological investigation to accompany the present study. Loan of museum materials is currently not possible, and we are unable to examine the type specimens of three previously synonymized taxa, A. typus (Ghana), A. bensonii (Liberia) and A. tschiloangensis (Cabinda, Angola), or investigate potential osteological (Parenti, 1981; Ghedotti, 2000; Costa, 2012) or morphometric variation among populations. Further investigation of potential phenotypic differentiation between the molecular lineages identified here is necessary prior to formalizing any taxonomic conclusions, and consequently, we must defer such actions to a future contribution.
The present study has, however, uncovered considerable structuring and genetic divergences between populations that are frequently 4–11 times higher than the traditionally employed sequence divergence heuristic threshold of 2–3% for teleostean conspecifics (e.g. Pereira et al., 2013; Decru et al., 2016; Iyiola et al., 2018; Arroyave et al., 2019) (Fig. 4b, Table 2). Here, we focus our discussion on the potential drivers of diversification and mechanisms that are likely to have shaped the contemporary distributions of lineages within this complex.
The onset of A. spilauchen diversification coincides with one of the main climatic shifts during the Neogene (Fig. 5). Initially, a period known as the Middle Miocene Optimum (between 16 and 14.8 mya) was warm and humid with high precipitation. This was followed by the Middle Miocene Climatic Transition (14.8–12.9 mya) which was much dryer leading to a significant drop in sea levels and increased aridification across the African continent (Flower & Kennett, 1994; Kender et al., 2009, 2014; Herold et al., 2011; Frigola et al., 2018). During the Middle Miocene Optimum, a moist and warm climate promoted the expansion of tropical vegetation, which covered most of the continent expanding even to high latitudes (Lovett, 1993; Maley, 1996; Plana, 2004; Stanley et al., 2005; Frigola et al., 2018). During this time, sea level rise likely engendered mangrove expansion throughout the Atlantic coast, and consequently potential expansion of an ancestral range for A. spilauchen. Our ancestral area analysis estimates an expansion from an area that corresponds to the present-day Ghanaian, Nigerian and Equatorial Guinean coastlines to adjacent Gabon to the south and Guinea to the west (Fig. 6). During the Middle Miocene Climatic Transition, this area experienced a marked contraction of mangrove habitats isolating Aplocheilichthys lineages in the westernmost coastal regions of Guinea and Ghana, and another in Nigeria, Equatorial Guinea and Gabon (Fig. 6).
Studies on the evolution of the Congo River drainage (Beadle, 1981; Burke, 1996; Giresse, 2005; Goudie, 2005; Stankiewicz & de Wit, 2006; Runge, 2007), deposition patterns in the Congo deep sea fan (Lavier et al, 2001; Leturmy et al., 2003; Lucazeau et al., 2003; Anka & Séranne, 2004; Anka et al., 2009; Savoye et al., 2009) and freshwater fish diversification in the basin (Goodier et al., 2011; Schwarzer et al., 2011; Alter et al., 2015, 2017; Arroyave et al., 2020; Stiassny & Alter, in press) reveal a complex geologic history for the most diverse river basin in Africa. In the western basin, sedimentary studies suggest a protracted history of shifting and intermittent outflow of the Congo River into the Atlantic during the Cenozoic. Although a final consensus has yet to be reached, a single high-energy capture event is now generally considered to have established the current Congo outlet to the Atlantic shortly after the Miocene-Pliocene transition. A Pliocene capture (5.3–2.6 mya) is supported by an increase in sediment deposition in the Congo fan and by tectonic activity along the Atlantic Rise during that time (Lavier et al., 2001; Leturmy et al., 2003; Lucazeau et al., 2003; Anka & Séranne, 2004; Anka et al., 2009; Savoye et al., 2009). High humidity during this period is also suggested by palynological data and probably these events facilitated a range expansion from Nigeria, Equatorial Guinea, and Gabon southwards to the regions in and around the newly formed lower Congo estuary at the border between the Democratic Republic of Congo and Angola (Fig. 6). Recent data on the extent of the Congo River plume and its influence on sea surface salinity and temperature over expansive coastal regions (Materia et al., 2012; Denamiel et al., 2013; Chao et al., 2015) highlights the importance of the origin of the present-day Congo River outlet for the expansion of mangroves throughout the region. During wet seasons, the Congo plume connects with the Niger River plume, and a lower salinity is recorded for the entire Gulf of Guinea up to and including the region around the Congo outlet.
West African aridification during the Pliocene (5.3–2.6 mya) is documented by both palynological and sediment data sampled from ocean drilling sites along the coast (Bonnefille et al., 1982; Leroy & Dupont, 1994; Vallé et al., 2017). Study of sedimentological sequences reveals a significant reduction in river discharge and an increase in wind borne grass pollen, both indicative of an arid climate. In addition, a concomitant reduction in mangrove pollen (Rhizophora spp.) suggests that aridification resulted in mangrove reduction and fragmentation. The late Pliocene aridification likely facilitated two vicariance events observed in our study: one between the Ghanaian and Guinean lineages and the other between the Nigerian and Equatorial Guinean lineages. Studies at sites close to Cape Blanc in Mauritania and just south of the mouth of the Senegal River found similar patterns, indicating an onset of aridification around 3.4 mya (Bonnefille et al., 1982; Leroy & Dupont, 1994). A similar study at a site near the Comoé River outlet, identified aridification in north western Africa between 3.5 and 2.9 mya (Vallé et al., 2017), a timing consistent with the proposed date for the split between Ghanaian and Guinean lineages (3.5 mya) estimated in the present study (Fig. 5)).
Other sites with detailed palynological information are near the Niger Delta, where a pronounced aridification with an increase in wind borne grass pollen and marked reduction of Rhizophora pollen is recorded between 2.7 and 2.0 mya (Durugbo et al., 2010; Adeonipekun et al., 2016). These dates are also consistent with the findings of the present study where the split between Nigerian and Equatorial Guinean lineages is estimated to have occurred around 2.5 mya (95% HPD 0.8–5.0 mya) (Fig. 5). Vicariance between Nigerian and Equatorial Guinean lineages may also be related to increased activity of the Cameroon Volcanic Line, a chain of mountains and volcanos that extends from the Cameroonian Highlands to the volcanic islands in the Gulf of Guinea (Deruelle et al., 1991; Marzoli et al., 2000; Burke, 2001). Bioko Island is located about 60 km from the mainland and experienced volcanic activity during the same period with volcanism of Cameroon and Manengouba mounts (around 3.0–1.0 mya) near the coast (Deruelle et al., 1991; Marzoli et al., 2000). Volcanism during low sea level would have had major impacts on water chemistry, temperature and coastal sedimentation directly impacting mangrove cover throughout the region. Recently, a comprehensive study of population connectivity of the mangrove, Rhizophora racemosa, around the Cameroonian and Equatorial Guinean coastline found high levels of genetic structuring related to the volcanic activity of Bioko Island and the Bioko-Cameroon land bridge formation (Ngeve et al., 2016). Here, we suggest that volcanism and the land bridge formation likely also affected A. spilauchen and resulted in the disruption of connectivity (gene flow) between Nigerian and Equatorial Guinean lineages.
The late Pleistocene-Holocene is known for considerable climate instability (Marius & Lucas, 1991; Scourse et al., 2005; Malounguila-Nganga et al., 2017; Maley et al., 2017; Molliex et al., 2019). Although our sampling of A. spilauchen from the southern extent of its range is limited, ancestral area reconstruction posits two additional vicariance events among haplotypes in the Kouilou, lower Congo and Angolan cluster during this timeframe (Fig. 6). Our results indicate a recent divergence and differentiation of the Kouilou haplotypes from the lower Congo and Angolan cluster. However, the low levels of genetic divergence detected, when compared to the significant genetic divergence between the other A. spilauchen populations, suggest the possibility of a continued gene flow between all of these populations (Fig. 4b, Table 2). A recent, and ongoing, connection is suggested by the influence of the Congo River freshwater plume, which is periodically carried south by the Angola current (Kopte et al., 2017), reaching beyond the Nyanga and Kwanza river ouflows (Denamiel et al., 2013). Similarly, an increase in mangrove vegetation persisting since the last deglaciation is evidenced by pollen data from the mouth of Congo (Scourse et al., 2005). Considerably denser population sampling of A. spilauchen throughout this southern region will be necessary to resolve this issue.
Conclusion
Despite relatively limited sampling, the applications of species delimitation, phylogeographic, and phylogenetic methods reveal a pattern of genetic differentiation within A. spilauchen that is concordant with a series of historical events recorded since the Middle Miocene. Our study highlights extremely low connectivity between most populations and a time-calibrated phylogeographic pattern that lends support to the novel hypothesis that a major driver of diversification within the lineage has been the shifting dynamics of coastal mangrove forest cover over time. We report, for the first time, a pattern of diversification within a lineage of brackish water fish that is concordant with the historical distribution of coastal mangroves forests, the predominant brackish water habitat of the focal species throughout its range.
From a conservation perspective, these results are of considerable significance since many brackish environments, particularly the mangrove forests, are highly threatened by coastal development and the exploration and extraction of hydrocarbons along the African Atlantic coastline (Alongi, 2015; Feka & Morrison, 2017). It has been estimated that mangroves are disappearing at a rate of 1–2% per year, suggesting the complete disappearance of these societally and biologically important ecosystems within the century (Alongi, 2015). The unexpected diversity observed in the A. spilauchen complex suggests that many of the other taxa that share a similar distribution associated with the same dynamic mangrove habitats may hide a significant, but currently undocumented, diversity under threat. Before formal taxonomic and nomenclatural changes can be undertaken, morphological analyses of the distinct populations (OTUs) identified herein are needed, and conservation assessments for each are critical as many of these potential new species are likely highly threatened by ongoing coastal development throughout the region.
References
Adeonipekun, P. A., M. Adebisi Sowunmi & K. Richards, 2016. A new Late Miocene to Pleistocene palynomorph zonation for the western offshore Niger Delta. Palynology. https://doi.org/10.1080/01916122.2015.1107652
Ahl, E., 1928. Beiträge zur Systematik der africanischen Zahnkarpfen. Zoologischer Anzeiger 79: 115–116.
Alongi, D. M., 2015. The Impact of Climate Change on Mangrove Forests. Current Climate Change Report 1: 30–39.
Alter, S. E., B. Brown & M. L. J. Stiassny, 2015. Molecular phylogenetics reveals convergent evolution in lower Congo River spiny eels. BMC Evolutionary Biology 15: 224.
Alter, S. E., J. Munshi-South & M. L. J. Stiassny, 2017. Genomewide SNP data reveal cryptic phylogeographicstructure and microallopatric divergence in a rapids-adapted clade of cichlids from the Congo River. Molecular Ecology 26: 1401–1419.
Anka, Z. & M. Séranne, 2004. Reconnaissance study of the ancient Zaire (Congo) deep-sea fan. (ZaiAngo Project). Marine Geology 209: 223–244.
Anka, Z., M. Séranne., M. Lopez., M. Scheck-Wenderoth & B. Savoye, 2009. The long-term evolution of the Congo deep-sea fan: A basin-wide view of the interaction between a giant submarine fan and a mature passive margin (ZaiAngo project). Tectonophysics 470: 42–56.
Arroyave, J., C. M. Martinez & M. L. J. Stiassny, 2019. DNA barcoding uncovers extensive cryptic diversity in the African long-fin tetra Bryconalestes longipinnis (Alestidae: Characiformes). Journal of Fish Biology 95: 379–392.
Arroyave, J., J. S. S. Denton & M. L. J. Stiassny, 2020. Pattern and timing of diversification in the African freshwater fish genus Distichodus (Characiformes: Distichodontidae). BMC Evolutionary Biology 20: 1–28.
Bartáková, V., M. Reichard., R. Blažek., M. Polačik & J. Bryja, 2015. Terrestrial fishes: rivers are barriers to gene flow in annual fishes from the African savannah. Journal of Biogeography 42: 1832–1844.
Beadle, L. C., 1981. The inland waters of tropical Africa. Longman, London.
Bennett, R. H., B. R. Ellender., T. Mäkinen., T. Miya., P. Pattrick., R. J. Wasserman., D. J. Woodford & O. L. F. Weyl, 2016. Ethical considerations for field research on fishes. Koedoe-African Protected Area Conservation and Science 58(1): 1–15.
Bleeker, P., 1863. Mémoire sur les poissons de la Côte de Guinée. Natuurkundige Verhandelingen der Hollandsche Maatschp voor Wetenschappen 18: 136.
Bonnefille, R., M. Rossignol-Strick & G. Riollet, 1982. Organic matter and palynology of DSDP site 367 Pliocene-Pleistocene cores off West Africa. Oceanologica Acta 5: 97–104.
Bragança, P. H. N. & W. J. E. M. Costa, 2019. Multigene fossil-calibrated analysis of the African lampeyes (Cyprinodontoidei: Procatopodidae) reveals an early Oligocene origin and Neogene diversification driven by palaeogeographic and palaeoclimatic events. Organisms, Diversity and Evolution 19: 1–18.
Burke, K., 1996. The African plate. South African Journal of Geology 99: 341–409.
Burke, K., 2001. Origin of the Cameroon Line of Volcano-Capped Swells. The Journal of Geology 109: 349–362.
Chao, Y., J. D. Farrara., G. Schumann., K. M. Andreadis & D. Moller, 2015. Sea surface salinity variability in response to the Congo River discharge. Continental Shelf Research 99: 35–45.
Costa, W. J. E. M., 2012. The caudal skeleton of extant and fossil cyprinodontiform fishes (Teleostei: Atherinomorpha): Comparative morphology and delimitation of phylogenetic characters. Vertebrate Zoology 62: 161–180.
Darriba, D., G. L. Taboada., R. Doallo & D. Posada, 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772.
Decru, E., T. Moelants., K. De Gelas., E. Vreven., E. Verheyen & J. Snoeks, 2016. Taxonomic challenges in freshwater fishes: A mismatch between morphology and DNA barcoding in fish of the north-eastern part of the Congo basin. Molecular Ecology Resources 16: 342–352.
Denamiel, C., W. P. Budgell & R. Toumi, 2013. The Congo River plume: Impact of the forcing on the far-field and near-field dynamics. Journal of Geophysical Research: Oceans 118: 964–989.
Deruelle, B., C. Moreau., C. Nkoumbou., R. Kambou., J. Lissom., E. Njonfang., R. T. Ghogumu & A. Nono, 1991. The Cameroon Line: a review. In Kampunzu A. B. & R. T. Lubala (eds), Magmatism in extension structure settings: the Phanerozoic African Plate. Springer, Berlin: 275–327.
Duke, N. C., 2017. Chapter 2: Mangrove Floristics and Biogeography Revisited: Further Deductions from Biodiversity Hot Spots, Ancestral Discontinuities, and Common Evolutionary Processes. In Rivera-Monroy V. H., S. Y. Lee., E. Kristensen & R. R. Twilley (eds), Mangrove Ecosystems: A Global Biogeographic Perspective: 17–52.
Durugbo, E. U., O. T. Ogundipe & O. K. Ulu, 2010. Palynological evidence of Pliocene-Pleistocene climatic variations from the western Niger Delta, Nigeria. International Journal of Botany 6: 351–370.
Drummond A. J., S. Y. W. Ho., M. J. Phillips & A. Rambaut, 2006. Relaxed phylogenetics and dating with confidence. PLoS Biology 4: e88.
Drummond, A. J., M. A. Suchard., D. Xie & A. Rambaut, 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29: 1969 –1973.
Duméril, A. H. A., 1861. Poissons de la côte occidentale d'Afrique. Archives du Muséum d'Histoire Naturelle10: 241 –268.
Feka, Z. N. & I. Morrison, 2017. Managing mangroves for coastal ecosystems change: A decade and beyond of conservation experiences and lessons for and from west-central Africa. Journal of Ecology and the Natural Environment 9: 99–123.
Flower, B. P. & J. P. Kennett, 1994. The middle Miocene climatic transition: East Antarctic ice sheet development, deep ocean circulation and global carbon cycling. Palaeogeography, Palaeoclimatology, Palaeoecology 108: 537–555.
Flugel, T. J., F. D. Eckardt & F. P. Cotterill, 2015. The present day drainage patterns of the Congo River system and their Neogene evolution. In De Wit, M. J., F. Guillocheau & M. C. J. De Wit (eds), Geology and Resource Potential of the Congo Basin. Springer Berlin Heidelberg: 315–337.
Folmer, O., M. Black., W. Hoeh., R. Lutz & R. Vrijenhoek, 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3: 294–299.
Frigola, A., M. Prange & M. Schulz, 2018. Boundary conditions for the Middle Miocene Climate Transition (MMCTv1.0). Geoscientific Model Development 11: 1607–1626.
Fujisawa, T. & T. G. Barraclough, 2013. Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: a revised method and evaluation on simulated datasets. Systematic Biology 62: 707–724.
Ghedotti, M. J., 1998. Phylogeny and classification of the Anablepidae (Teleostei: Cyprinodontiformes). In Malabarba, L. R., R. E. Reis., R. P. Vari., Z. M. S. Lucena & C. A. S. Lucena (eds), Phylogeny and classification of Neotropical fishes.Edipucrs, Porto Alegre: 561–582.
Ghedotti, M. J., 2000. Phylogenetic analysis and taxonomy of the poecilioid fishes (Teleostei: Cyprinodontiformes). Zoological Journal of the Linnean Society 130: 1–53.
Giresse, P., 2005. Mesozoic−Cenozoic history of the Congo Basin. Journal of African Earth Sciences 43: 301–315.
Goodier, S. A. M., F. P. D. Cotterill., C. O'Ryan., P. H. Skelton & M. J. de Wit, 2011. Cryptic diversity of African Tigerfish (genus Hydrocynus) reveals palaeogeographic signatures of linked neogene geotectonic events. PLoS One 6: e28775.
Goudie, A. S., 2005. The drainage of Africa since Cretaceous. Geomorphology 67: 437–456.
Günther, A., 1866. Catalogue of the fishes in the British Museum, Volume 6. The British Museum (Natural History): London.
Herbert, P. D. N., A. Cywinska., S. L. Ball & J. R. De Waard, 2003. Biological identifications through DNA barcodes. In: Proceedings of the Royal Society of London. Series B 270: 313–321.
Herold, N., M. Huber & R. D. Müller, 2011. Modeling the miocene climatic optimum. Part I: Land and atmosphere. American Meteorological Society 24: 6353–6372.
Ho, S. Y. W., 2007. Calibrating molecular estimates of substitution rates and divergence times in birds. Journal of Avian Biology 38: 409–414.
Ho, S. Y. W. & M. J. Phillips, 2009. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Systematic Biology 58: 367–380.
Huber, J. H., 1982. Cyprinodontidés recoltés en Côte d’Ivoire (1974-1978). Cybium 6: 49–73.
Huber, J. H., 1999. Updates to the phylogeny and systematics of the African lampeye schooling cyprinodonts (Cyprinodontiformes: Aplocheilichthyinae). Cybium 23: 53–77.
Iyiola, O. A., L. M. Nneji., M. K. Mustapha., C. G. Nzeh., O. S. Oladipo., I. C. Nneji., A. O. Akeyoyin., C. D. Nwani., O. A. Ugwumba., A. A. A. Ugwumba., E. O. Faturoti., Y. Y. Wang., J. Chen., W. Z. Wang & A. C. Adeola, 2018. DNA barcoding of economically important freshwater fish species from north-central Nigeria uncovers cryptic diversity. Ecology and Evolution 8: 6932–6951.
Kathiresan, K. & B. L. Bingham, 2001. Biology of Mangroves and Mangrove Ecosystems. Advances in Marine Biology 40: 81–251.
Kender, S., V. L. Peck., R. W. Jones & M. A. Kaminski, 2009. Middle Miocene oxygen minimum zone expansion offshore West Africa: Evidence for global cooling precursor events. Geology 37: 699–702.
Kender, S., J. Yu & V. L. Peck, 2014. Deep ocean carbonate ion increase during mid Miocene CO2 decline. Scientific Reports 4: 4187.
Kimura, M., 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120.
Kopte, R., P. Brandt., M. Dengler., P. C. M. Tchipalanga., M. Macuéria & M. Ostrowski, 2017. The Angola Current: Flow and hydrographic characteristics as observed at 11oS. Journal of Geophysical Research: Oceans 122: 1177–1189.
Kruskal, J. B., 1956. On the shortest spanning subtree of a graph and the traveling salesman problem. Proceedings of the American Mathematical Society 7: 48–50.
Kumar, S., G. Stecher & K. Tamura, 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874.
Lanfear, R., P. B. Frandsen., A. M. Wright., T. Senfeld & B. Calcott, 2016. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773.
Lavier, L. L., M. S. Steckler & F. Brigaud, 2001. Climatic and tectonic control on the Cenozoic evolution of the West African margin. Marine Geology 178: 63–80.
Leigh, J. W. & D. Bryant, 2015. Popart: full‐feature software for haplotype network construction. Methods in Ecology and Evolution 6: 1110–1116.
Leroy, S. & L. Dupont, 1994. Development of vegetation and continental aridity in northwestern Africa during the Late Pliocene: the pollen record of ODP site 658. Palaeogeography, Palaeoclimatology, Palaeoecology 109: 295–316.
Leturmy, P., F. Lucazeau & F. Brigaud, 2003. Dynamic interactions between the Gulf of Guinea passive margin and the Congo River drainage basin: 1. Morphology and mass balance. Journal of Geophysical Research: Solid Earth 108: 2383.
Lovett, J. C., 1993. Eastern Arc moist forest flora. In Lovett, J. C. & S. K. Wasser (eds), Biography and ecology of the rain forests of eastern Africa. Cambridge University Press, Cambridge: 33–35.
Lucazeau, F., F. Brigaud & P. Leturmy, 2003. Dynamic interactions between the Gulf of Guinea passive margin and the Congo River drainage basin: 2. Isostasy and uplift. Journal of Geophysical Research: Solid Earth 108: 2384.
Luther, D. A. & R. Greenberg, 2009. Mangroves: A Global Perspective on the Evolution and Conservation of Their Terrestrial Vertebrates. BioScience 59: 602–612.
Malounguila-Nganga, D., P. Giresse., M. Boussafir & T. Miyouna, 2017. Late Holocene swampy forest of Loango Bay (Congo). Sedimentary environments and organic matter deposition. Journal of African Earth Sciences 134: 419–434.
Maddison, W. P. & D. R. Maddison, 2019. Mesquite: a modular system for evolutionary analysis. Version 3.61 http://www.mesquiteproject.org.
Maley, J., 1996. The African rainforest: main characteristics of changes in vegetation and climate from the Upper−Cretaceous to the Quaternary. Proceedings of the Royal Society of Edimburgh Section B 104: 31–73.
Maley, J., C. Doumengec., P. Giressed., G. Mahéa., N. Philipponf., W. Hubau., M. O. Lokondah., J. M. Tshibambag & A. Chepstow-Lustyk, 2017. Late Holocene forest contraction and fragmentation in central Africa. Quaternary Research 1–17.
Marius, C. & J. Lucas, 1991. Holocene Mangrove Swamps of West Africa Sedimentology and Soils. Journal of African Earth Sciences 12: 41–54.
Marzoli, A., E. M. Piccirillo., P. Renne., G. Bellieni., M. Lacumin., N. B. Nyobe & A. T. Tongwa, 2000. The Cameroon Volcanic Line revisited: petrogenesis of continental basaltic magmas from lithospheric and asthenospheric mantle sources. Journal of Petrology 41: 87–109.
Materia, S., S. Gualdi., A. Navarra & L. Terra, 2012. The effect of Congo River freshwater discharge on Eastern Equatorial Atlantic climate variability. Climate Dynamics 39: 2109–2125.
Miller, M. A., W. Pfeiffer & T. Schwartz, 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Gateway computing environments workshop (GCE). IEEE, Red Hook, NY: 1–8.
Molliex, S., A. J. Kettner., D. Laurent., L. Droz., T. Marsset., A. Laraque., M. Rabineau & G. D. Moukandi N’Kaya, 2019. Simulating sediment supply from the Congo watershed over the last 155 ka. Quaternary Science Reviews 203: 38–55.
Ngeve, M. N., T. Van der Stocken., D. Menemenlis., N. Koedam & L. Triest, 2016. Contrasting Effects of Historical Sea Level Rise and Contemporary Ocean Currents on Regional Gene Flow of Rhizophora racemosa in Eastern Atlantic Mangroves. PLoS ONE 11:e0150950.
Okyere, I., 2012. Some ambient environmental conditions, food and reproductive habits of the banded lampeye killifish Aplocheilichthys spilauchen in the Kakum estuary wetland, Ghana. Environmental Biology of Fishes 94: 639–647.
Parenti, L. R., 1981. A phylogenetic and biogeographical analysis of cyprinodontiform fishes (Teleostei: Atherinomorpha). Bulletin of the American Museum of Natural History 168: 335–557.
Pereira, L. H., R. Hanner., F. Foresti & C. Oliveira, 2013. Can DNA barcoding accurately discriminate megadiverse neotropical freshwater fish fauna? BMC Genetics 14: 20.
Peters, W. C. H., 1864. Berichte uber einige neue Saugetiere, Amphibien und Fische. Monatsberichte der Akademie der Wissenschaffen in Berlin 1864: 381–399.
Plana, V., 2004. Mechanisms and tempo of evolution in the African Guineo−Congolian rainforest. Philosophical Transactions of the Royal Society London B Biological Sciences 359: 1585–1594.
Puillandre, N., A. Lambert., S. Brouillet & G. Achaz, 2012. ABGD, automatic barcode gap discovery for primary species delimitation. Molecular Ecology 21: 1864–1877.
Rambaut, A., M. A. Suchard., D. Xie & A. J. Drummond, 2014. Tracer v1.6. Retrieved from http://beast.bio.ed.ac.uk/Tracer.
Ronquist, F., M. Teslenko., P. van der Mark., D. Ayres., A. Darling., S. Höhna., B. Larget., L. Liu., M. A. Suchard & J. P. Huelsenbeck, 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539 –542.
Runge, J., 2007. The Congo River, Central Africa. In Gupta, A. (ed.), Geomorphology and Management. Wiley, Hoboken, NJ: 293–309.
Saenger, P. & M. E. Bellan, 1995. The mangrove vegetation of the Atlantic coast of Africa: A review. Universitè de Toulouse Press: Toulouse.
Savoye, B., N. Babonneau., B. Dennielou & M. Bez, 2009. Geological overview of the Angola–Congo margin, the Congo deep-sea fan and its submarine Valléys. Deep Sea Research Part II: Topical Studies in Oceanography 56: 2169-2182.
Schwarzer, J., B. Misof., S. N. Ifuta & U. K. Schliewen, 2011. Time and origin of cichlid colonization of the lower Congo rapids. PLoS One 6: e22380.
Scourse, J., F. Marret., G. J. M. Versteegh., J. H. Fred Jansen., E. Schefß & J. van der Plicht 2005. High-resolution last deglaciation record from the Congo fan reveals significance of mangrove pollen and biomarkers as indicators of shelf transgression. Quaternary Research 64: 57-69.
Stadler, T., 2009. On incomplete sampling under birth–death models and connections to the sampling-based coalescent. Journal of Theoretical Biology 261: 58–66.
Stankiewicz, J. & M. J. de Wit, 2006. A proposed drainage evolution model for Central Africa – did the Congo flow east? Journal of African Earth Sciences 44: 75–84.
Stanley, W. T., M. A. Rogers & R. Hutterer, 2005. A new species of Congosorex from the Eastern Arc Mountains, Tanzania, with significant biogeographical implications. Journal of Zoology 265: 269–280.
Stiassny, M. L. J., & S. E. Alter, in press. Evolution in the fast lane: diversity, ecology, and speciation of cichlids in the lower Congo River. In Abate, M. E. & D. L. G. Noakes (eds), The Behavior, Ecology, and Evolution of Cichlid Fishes. Fish and Fisheries Series. Springer, New York.
Sunnucks, P., P. R. England., A. C. Taylor & D. F. Hales, 1996. Microsatellite and chromosome evolution of parthenogenetic Sitobion aphids in Australia. Genetics 144: 747–756.
Thomaz, A. T., L. R. Malabarba., S. L. Bonatto & L. L. Knowles, 2015. Testing the effect of palaeodrainages versus habitat stability on genetic divergence in riverine systems: study of a Neotropical fish of the Brazilian coastal Atlantic Forest. Journal of Biogeography 42: 2389–2401.
Thomaz, A. T. & L. L. Knowles, 2018. Flowing into the unknown: inferred paleodrainages for studying the ichthyofauna of Brazilian coastal rivers. Neotropical Ichthyology 16: e180019.
Vallé, F., T. Westerhold & L. M. Dupont, 2017. Orbital-driven environmental changes recorded at ODP Site 959 (eastern equatorial Atlantic) from the Late Miocene to the Early Pleistocene. International Journal of Earth Sciences 106: 1161–1174.
Ward, R. D., 2009. DNA barcode divergence among species and genera of birds and fishes. Molecular Ecology Resources 9: 1077–1085.
Wildekamp, R. H., 1995. A world of killies, atlas of the oviparous cyprinodontiform fishes of the world (Vol. 2). American Killifish Association: Mishawaka.
Wildekamp, R. H., R. Romand & J. J. Scheel, 1986. Cyprinodontidae. In Daget, J., J. P. Gosse & T. van den Audenaerde (eds), Checklist of the freshwater fishes of Africa 2 (CLOFFA2). ISNB, MRAC, ORSTOM, Brussels, Tervuren, Paris: 165–276.
Wright, J. M., 1986. The ecology of fish occurring in shallow water creeks of a Nigerian mangrove swamp. Journal of Fish Biology 29: 431–441.
Xia, X. H., 2013. Dambe5: a comprehensive software package for data analysis in molecular biology and evolution. Molecular Biology and Evolution 30: 1720–1728.
Xia, X. H., Z. Xie., M. Salemi., L. Chen & Y. Wang, 2003. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution 26: 1–7.
Yu, Y., A. J. Harris & X. J. He, 2010. S-DIVA (statistical dispersal-vicariance analysis): A tool for inferring biogeographic histories. Molecular Phylogenetics and Evolution 56: 848–850.
Yu, Y., A. J. Harris., C. Blair & X. He, 2015. RASP (reconstruct ancestral state in phylogenies): A tool for historical biogeography. Molecular Phylogenetics and Evolution 87: 46–49.
Zhang, J., P. Kapli., P. Pavlidis & A. Stamatakis, 2013. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29: 2869–2876.
Zwickl, D. J., 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. dissertation, The University of Texas at Austin.
Acknowledgments
Our thanks to J. Snoeks and M. Parrent (RMCA), T. Vigliotta, R. Arrindell and C. Lewis (AMNH), W. Costa (UFRJ), B. Sidlauskas and P. Konstantinidis (OS), J. Williams and D. Pitassy (USNM), I. Okyere (University of Cape Coast, Ghana), J. Cutler (University of California, Santa Cruz, USA) and J. Hervé Mve Beh (Libreville, Gabon) for the donation or loan of specimens and tissue samples. We thank A. Katz (UFRJ) R. Bills (SAIAB) and K. Bernotas (AMNH) for photographing preserved specimens, and C. Aubin (Périgueux, France), L. Chirio (Brazzaville, Republic of the Congo), L. Kent (Seattle, USA), P. Venstermans (Zwijndrecht, Belgium), and R.B. Tate (Witrivier, South Africa) for providing pictures of live specimens. We gratefully acknowledge P. Amorim and J. Mattos (UFRJ) and T. Ntokoane (SAIAB) for assistance in the Molecular Laboratory, and for the use of equipment provided by the UFRJ Ichthyology Laboratory and the NRF-SAIAB Molecular Genetic Laboratory. Partial funding for this project was provided by the National Geographic Society (#WW-055R-17) and Randolph-Macon College, and the Axelrod Curatorship of the American Museum of Natural History.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by P. Bragança and R. Schmidt. Figures were prepared by M. Stiassny and P. Bragança. The first draft of the manuscript was written by P. Bragança, and all authors commented and provided changes on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Additional information
Handling editor: Christian Sturmbauer
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bragança, P.H.N., Van der Zee, J., Chakona, A. et al. Following the Mangroves: diversification in the banded lampeye Aplocheilichthys spilauchen (Duméril, 1861) (Cyprinodontiformes: Procatopodidae) along the Atlantic coast of Africa. Hydrobiologia 848, 1433–1453 (2021). https://doi.org/10.1007/s10750-020-04497-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-020-04497-3