Abstract
The ubiquitous and species rich genus Ulva comprises entities of green macroalgae with variable morphologies. Ulva species are important from ecological and economic perspectives, but their identification is often problematic. Current knowledge on Ulva diversity has focused mainly on foliose individuals of temperate regions, but genetic and morphological data on tubular species are often insufficient and the species richness is ambiguous due to the lack of molecularly identified type vouchers. Together with a previous study, our study demonstrates that due to the crypticity of tubular entities of the genus Ulva present in the Atlantic-Baltic Sea transect, certain species remained undetected until recently whereas molecular evidence of other historically identified species is missing. An entity which appears to be a relatively frequent species in the Atlantic-Baltic Sea transect and which was probably mis-identified with other species in the past is here described as Ulva capillata sp. nov.. The description is based on molecular identification using tufA and rbcL sequences, and by comparing the species´ phylogenetic relationships, distribution and range margins in the Atlantic-Baltic Sea transect, as well as on morpho-anatomical characters, and early ontogenetic development. By comparisons with closely related and potentially morphologically overlapping species concepts we were able to identify the uniqueness of U. capillata. Therefore, the description of U. capillata as a new species within the genus Ulva is supported by a combination of molecular, morphological, and ontogentic evidence which confirm their uniqueness in comparison to other species concepts.
Furthermore, our results strongly emphasize the importance and necessity to molecularly investigate especially tubular historic type vouchers within the genus Ulva to facilitate a clear species identification to omit continuing with taxonomic confusion and ongoing misapplication of names of e.g. cryptic species concepts within this important green algal genus.
Similar content being viewed by others
Introduction
Green algae of the genus Ulva (Linnaeus 1753) are ubiquitous green seaweeds that inhabit a range of salinity conditions – from fully marine over brackish to freshwater – which makes it the most cosmopolitan genus of the order Ulvales (Lagourgue et al. 2022). Ulva species are considered nitrophilic organisms, as they can be indicative of eutrophic environments (Kraft et al. 2010) and generally have high metabolic growth rates (Rosenberg and Ramus 1984; Teichberg et al. 2010). Due to their ability to proliferate and form blooms (also called green tides) (Blomster et al. 1998; Smetacek and Zingone 2013; Cai et al. 2021), some Ulva species have attracted interest both from state agencies and policy makers (e.g. driven by the EU´s Water Framework Directive 2000/60/EG) to support biodiversity monitorings, as well as from the emerging aquaculture sector as suitable crop strains (Bolton et al. 2009; Carl et al. 2014; Califano et al. 2020; Olsson et al. 2020; Steinhagen et al. 2021; 2022a; b). However, not all species share the same traits, and most Ulva species are important primary producers in healthy ecosystems and may not be suitable for cultivation. Therefore, in order to detect nuisance species or ecotypes, enable monitoring and observation of invasive species, and to evaluate the suitability for commercial cultivation, correct species identification should be a central part of any research or industry involving Ulva.
The species rich genus Ulva encompasses > 550 species names, including several described subspecies, varieties, and forms (Guiry and Guiry 2022). During the last decade, several new species have been described (Chen et al. 2015; Phillips et al. 2016; Krupnik et al. 2018; Lagourgue et al. 2022) and allegedly well-defined species concepts have been taxonomically revised (Hughey et al. 2019, 2022; Steinhagen et al. 2019a, b; Fort et al. 2022; Tran et al. 2022). Today, 85 species are currently regarded taxonomically valid and > 70 species are declared with an uncertain taxonomic status (Guiry and Guiry 2022). The recent rise of taxonomic changes is associated with the occurrence of several cryptic species within the genus Ulva that have been detected by the availability of genetic methods, such as DNA barcoding (e.g. Hayden et al. 2003; Steinhagen et al. 2019a, b, c). Genetic markers most suitable for species identification and delimitation within the genus Ulva are tufA, rbcL, and ITS (Kraft et al. 2010; Tran et al. 2022). However, a re-occurring problem of DNA barcoding studies (which widely rely on well annotated and curated genetic databases) is the absence of suitable reference sequences of vouchers of lecto- and holotypes (Tran et al. 2022). Assigning species names to genetic sequences and reliably identifying organisms that have a highly cryptic diversity (such as the genus Ulva) requires access to, and genetic information of, the respective type vouchers of the species.
Under the genus Ulva, foliose forms (widely known as sea lettuces) and tubular forms (formerly allocated to the genus Enteromorpha but placed in synonymy with the genus Ulva based on genetic data) (Hayden et al. 2003) are grouped together. Whereas recent efforts have been made to molecularly identify foliose type specimens (Steinhagen et al. 2019a; Fort et al. 2021, 2022; Hughey et al. 2022; Tran et al. 2022), information for tubular Ulva species is limited to recently described species (e.g. Chen et al. 2015; Lagourgue et al. 2022). Sequencing of type material of tubular Ulva species is necessary as some species have variable and sometimes aberrant morphologies depending on abiotic environmental conditions (e.g. Bliding 1963; Blomster et al. 2002; Brodie et al. 2007; Steinhagen et al. 2019a, b, c, 2022c), which eventually leads to wrong species descriptions, false identifications, and the appearance of long persisting mis-naming in the literature.
The aim of the present study was to constitute detailed assessments of the molecular, morphological, and ontogenetic traits of a relatively frequently occuring but yet undescribed Ulva species that was discovered during a large-scale field survey during the years 2018–2022 in the Atlantic-Baltic Sea transect (see also Steinhagen et al. 2022c). By combining tufA and rbcL sequencing with morphological and ontogenetic observations, as well as by including detailed literature searches of the green algal biodiversity of the area, we were able to describe this entity as Ulva capillata sp. nov. and shed light onto the identity of a frequent species of the NE Atlantic and SW Baltic Sea. In addition, our study highlights past difficulties in distinguishing and delimiting tubular Ulva species in the area.
Materials and methods
Study area, field collection and sample preparation
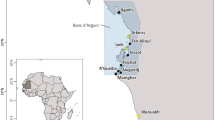
During a large-scale assessment of the Ulva biodiversity in the Atlantic- Baltic Sea transect over 287 sampling sites – covering the full salinity gradient in the Baltic Sea (from fully marine conditions to fresh water state (Steinhagen et al. 2022c)) and including sampling sites in Sweden (121), Denmark (66), Germany (54), Norway (26), and Finland (20) – were repeatedly visited in the years 2018–2022 (see also Steinhagen et al. 2022c). A variety of habitats (including rockpools, harbours, marine national parks, estuaries, fjords, drain channels, as well as exposed and sheltered coastal areas) were included in our sampling to reflect the different ecosystems present in the Atlantic-Baltisc Sea transect. Furthermore, different substrates (organic and inorganic, natural and artificial) of the attached thalli were included and drift populations were sampled as well. Generally, algae collections were conducted to a depth of ~ 1.5 m below mean sea level in the supra- and midlittoral zones using waders. Additionally, samplings of the mid- and infralittoral zones of chosen sites were conducted via snorkelling. Salinty (PSU), water temperature (°C) and oxygen levels (mg L−1) were measured using a WTW portable conductivity meter (Xylem Analytics, Germany) at most of the sites (see also Steinhagen et al. 2022c). An unidentified Ulva sp. (Ulva sp. 2) was encountered at 15 of the sampling sites (Fig. 1 and Table 1). Chosen sites were re-visited in 2022, to verify the presence of populations and obtain material for cultivation and ontogentic examinations. Specimens were collected, placed into sealed plastic bags, and stored on ice until further processing in the lab. Most samples were preserved as herbarium vouchers and lodged in the Herbarium of the University of Gothenburg [GB] (for voucher numbers see Table 1). A subsample was divided, with part of it stored at 4 °C or -20 °C for subsequent morphological observation and part of it stored in a microreaction tube at -80 °C for genomic DNA extraction and subsequent DNA barcoding.
Molecular biology and phylogenetic analysis
Genomic DNA was isolated from the lyophilized algal tissue of 21 specimens of Ulva sp. 2 with an Invisorb Spin Plant Mini Kit (Stratec, Germany) following the manufacturer´s protocol. Extracted DNA was stored at -80 °C and used for amplification of the rbcL and tufA genes to identify specimens by DNA barcoding. PCR amplifications of the rbcL gene used the primer pairs rbcLstart and R750, as well as F650 and rbcLend (Shimada et al. 2003). The PCR reactions were performed as follows: 94 °C for 1 min; 35 cycles at 94 °C for 30 s, at 56.3 °C for 30 s, and at 72 °C for 1 min; and a final extension step at 72 °C for 7 min. PCR amplification of the tufA gene followed the detailed description of Steinhagen et al. (2019a). PCR products were purified using the QIAquick PCR Purification Kit (Quiagen Germany). Sanger sequencing of the purified amplicons was provided by Eurofins Genomics (Konstanz, Germany). Forward and reverse sequence reads of the respective genes were assembled to produce contigs in Sequencher (v. 4.1.4, Gene Codes Corporation, USA) and a multiple sequence alignment was constructed for each gene region using MAFFT (Katoh et al. 2002). All sequences obtained in this study are publicly available in GenBank (for GenBank accession numbers see Table 1).
By applying the BLAST function in GenBank, first identification efforts based on the specimens’ rbcL and tufA sequences were made and closest related sequences were assessed. For resolving species identities peer-reviewed and annotated reference sequences downloaded from GenBank were included in subsequently generated sequence alignments. Emphasis was especially put on including reference sequences of holotype and lectotype material (Fig. 2) and all available sequences of tufA and rbcL of Ulva type material were included in downstream phylogenetic analyses.
Comparative maximum likelihood phylograms of tufA and rbcL sequences from taxa of Ulva, including specimens of Ulva sp. 2 from the Atlantic-Baltic Sea transect. The grey shaded boxes indicate clades of Ulva sp. 2, later described as Ulva capillata sp. nov. that were present in the study area and connects the respective clades in both phylogenetic analyses for direct comparison. Numbers at nodes indicate bootstrap values. Poorly supported nodes (< 70% bootstrap support) are not labelled. Branch lengths are proportional to sequence divergence
The rbcL and tufA sequences were analysed as separate datasets. Newly generated sequences were aligned with reference sequences downloaded from GenBank and used for further phylogenetic analysis. The models that best fit our data were found under the Akaike Information Criterion by employing MrModeltest software version v. 2.2. (Nylander 2004). For both datasets the optimal substitution model was determined and found to be GTR + Γ + I. Maximum-likelihood (ML) analyses were then carried out using RAxML version 8 (Stamatakis 2014), employing the chosen substitution model with 1000 bootstrap replicates for each alignment.
Morphological analysis
Morphological pre-identification of every tissue sample was based on typical morpho-anatomical characters (e.g. overall thallus morphology, cell form, cell arrangement, number of pyrenoids per cell, orientation of the chloroplst) and observations were based on original diagnoses and identification criteria of identification keys and previous studies of the respective area (Kylin 1949; Bliding 1963; Rueness 1977; Koeman and Van den Hoek 1981; 1982a; b; 1984; Hoeksema and Van den Hoek 1983; Blomster et al. 2002; Steinhagen et al 2019a, b). Lugol’s solution was used to stain starch containing compartments, such as pyrenoids. Macromorphological characters were observed on fresh and frozen material with a stereomicroscope and micromorphological characters were observed with a microscope fitted with a camera to capture photomicrographs.
Cultivation and early ontogenetic observations. Complete thalli of selected mature Ulva sp. 2 specimens were washed thoroughly and repeatedly with sterile seawater to remove dirt and adhering impurities and were isolated into cultures. Clean thalli were transferred into 150 × 20 mm petri dishes that were covered with microscopic slides on their bottom and were incubated in sterile artificial seawater adjusted to the salinity of the respective sites at 15 °C under a photon flux density of 90 µmol photons m−2 s−1 and a 16:8 h light: dark photo regime. To avoid nutrient depletion, Provasoli Enriched Seawater (Provasoli 1968) was added to the cultures. A bi-weekly water change and refreshment of medium was performed. To prevent the growth of diatoms, 1 mg L−1 GeO2 was added. The thalli were examined daily for sporulation events. After sporulation had taken place, the swarmers settled on the microscopic slides arranged on the bottom of the culture dishes and adult thalli were removed. The spore development was observed via light microscopy and photographically documented.
Results
Phylogeny
The phylogenetic analyses performed on datasets of the rbcL and tufA markers resulted in comparable and almost identical results and tree topologies and displayed equivalent evolutionary relationships of both investigated marker genes (Fig. 2). The rbcL alignment consisted of a total of 1313 positions, whereas the tufA gene dataset was 771 basepairs long.
The node separating the genus Ulva from the closely related outgroup taxon Umbraulva received full bootstrap support for both marker genes and unequivocably confirmed the taxonomic position of Ulva sp. 2 within the genus Ulva (similar results were obtained with more outgroup taxa including e.g., Blidingia, Kornmannia, and Monostroma – data not shown). All sequences identified as Ulva sp. 2 clustered closely together, forming a well delimited claded with full bootstrap support. The clades delimiting Ulva sp. 2 showed low intraspecific genetic variability for both marker genes (tufA: 0—0.12%, distance to next relative U. iliohaha 6.7–6.8%; rbcL: 0 – 0.15%, distance to next relative U. iliohaha 2.66—3.15%) and could not be resolved with reference sequences for the tufA gene. However, the rbcL phylogram revealed that sequences identified as Ulva ralfsii (Harvey) Le Jolis from New Zealand (rbcL GenBank accession numbers: MW250804, MW250805, MW250816) clustered within the Ulva sp. 2 clade (Fig. 2).
Both the tufA and rbcL data sets revealed the foliose Hawaiian species Ulva iliohaha H.L.Spalding & A.R.Sherwood (tufA GenBank accession number: KT932976; rbcL GenBank accession number: KT932995) as the most closest related to Ulva sp. 2, followed by a clade of newly described, tubular species from New Caledonia (e.g. Ulva batuffolosa Lagourgue & Payri, Ulva planiramosa Lagourgue, Ulva scolopendra Lagourgue & Payri) (Fig. 2).
In the following section the previously unidentified but apparently common entity Ulva sp. 2 will be described in more detail. Macro- and micromorphological observations, including respective ontogenetic features, as well as ecological and distribution information, are presented:
Ulva sp. 2
Habitat and distribution
Specimens of Ulva sp. 2 were found in the Atlantic, Skagerrak and Kattegat region of the Atlantic-Baltic Sea transect, and the southernmost distribution limit was reached after passing the Danish Straits in the Flensborg Fjord. It was encountered in habitats with salinities ranging from 31.3 to 14.4 PSU within this study (Fig. 1 and Table 1). The tubular and filigree thalli of Ulva sp. 2 were often found entangled as dense drifting mats (Fig. 3A) which were also encountered as patchy mats on the coast when beached, or they were found attached to different artificial and natural hard substrates – e.g. on ropes (Fig. 3B), epiphytic on other macrophytes (Fig. 3C), on concrete (Fig. 3D), or on mussle shells (Fig. 3E). Notably, populations and single individuals of Ulva sp. 2 were found in various habitats, ranging from open shore coatslines with strong wave action, to marinas and strongly trafficked harbours, to the pristine and shallow waters of the Kosterhavet National Park (Fig. 1 and Table 1). Attached populations of Ulva sp. 2 were predominantly found in the upper- to mid-littoral zone (Fig. 3C–E), seldomly in the infralittoral zone, whereas proliferating and drifting populations were more frequently found in shallow bays and enclosed bights and top layers of the drifting biomass were air exposed (Fig. 3A). Populations of Ulva sp. 2 were predominantly observed during summer (June–August) and only few individuals were encountered during winter or early spring.
Morphology of Ulva capillata sp. nov.. (A) Type locality of U. capillata at Båleröd in a sheltered bay of the Skagerrak region, Sweden. The thalli were either free floating (A) and entangled in drifting mats (i), or growing as tufts attached to artificial substrates such as ropes (B) or growing on stone or epiphytic on (C) Fucus. Furthermore, individuals in the Atlantic-Baltic Sea region were found attached to (D) concrete and (E) mussle shells. The tubular, branched individuals were often encountered to grow as (F–G) tufts or as (H–I) single individuals. Typically, the cells formed clear, longitudinal rows (J–L) which were observed to proceed also in the (J) side-branches. The cells were quadratic, rectangular, often polygonal with sharp angular corners and the chloroplast was parietal or filled the cell and contained 2–5 (rarely 1, or 6) randomly located pyrenoids (J–M). For a better resolution of the pyrenoids they were stained wih Lugol´s solution (M)
Morphology
The distinct, tubular, and mostly delicate and branched thalli were either compressed or inflated and reached 0.1 – 2.7 mm in width and 20 – 25 cm in length (rarely wider or taller) (Fig. 3F–H). The width of the thallus often increased as it proceeded from the rhizoidal zone to the tip. Branches were observed at around 70% of the investigated individuals (n = 95) and were uni- to multiseriate and present across the whole thallus (Fig. 3F–J). Unbranched individuals were mainly encountered drifting and were usually lacking a rhizoidal zone. All attached individuals exhibited branches. Often, multiple individuals shared the same thallus base and rhizoidal zone (Fig. 3F–G), resulting in a bushy appearance of entangled tufts (Fig. 3D–E) that could be perceived as dense clumps of Ulva sp. 2 (Fig. 3B–E). Branches and microscopic appendices were either sharp- or blunt-ended. Cells formed clear, longitudinal rows (Fig. 3J–L), which only seldomly blurred in broader thallus areas of the apical region and were even maintained in branch-axils and teeing branches (Fig. 3J). Individuals with unordered cell arrangements were observed infrequently. The cells were quadratic, rectangular, or polygonal with sharp angular, seldomly rounded corners and 9.3–26.4 (45) µm long and 4.8–20 µm broad (Fig. 3K–M). The chloroplast was often parietal (Fig. 3K) or filled the cell (Fig. 3L) and frequently the chloroplast was located at the same side of the cell within the single cell rows (Fig. 3K). The cells contained 2–5 (rarely 1 or 6) randomly located pyrenoid(s) (Fig. 3M).
Ontogeny
Only the ontogentic development of gametes of Ulva sp. 2 will be discussed, as no sporophytic individuals were obtained from wild stocks. After thoroughly washing the mature thalli of Ulva sp. 2 and placing them in culture vessels with sterile seawater, gametangia started to form within 1–3 days within the single cells (Fig. 4A–B). Three days after first observations of gametogenesis, swarmers were released from the gametangia (Fig. 4C–D). The pyriform, biflagellate gametes had a length ranging from 3.5–8.6 µm and a width ranging from 2.2–6.9 µm containing a visible eyespot (Fig. 4E). The motile and positive phototactic gametes attached (Fig. 4F) and started to secrete a cell wall (Fig. 4G). After the gamete settled, a germination-tube began to form (Fig. 4H–J). By distal, mitotic cell divisions a monostromatic, filament began to form (Fig. 4L). Subsequently, lateral cell division was observed and and an erect tube formed (Fig. 4M–N). First branching was observed after a minimum of c.a. 14 days (Fig. 4O).
Ontogenetic development of clonal gametophytes of Ulva capillata. After induction of gametogenesis, ganetagia started to form within 1–3 days within the single cells (A–B) and swarmer release was observed at day three (C–D). The pyriform, biflagellate gametes (E) contained an eyespot and eventually rounded off (F) and started to secrete a cell wall (G). After settlement a germination-tube began to form (H–J) and a monostromatic, filament began to form (L). Subsequently an erect tube formed (M–N) and side-branches were observed after about two weeks (O)
Comparison with other Ulva spp. concepts
Ulva sp. 2 shows a distinct morphology in most of the investigated individuals that can be distinguished from most existing Ulva type descriptions. In a large survey of the Ulva biodiversity conducted throughout the full Atlantic-Baltic Sea gradient, several encountered Ulva entities could be resolved with GenBank reference sequences (see also Steinhagen et al. 2022c). However, some species that were predicted to occur in the evaluated area, could not be verified with molecular data and, furthermore, for some “literature-predicted species”, no sequenced reference material was available to facilitate molecular identification. In a literature review focusing on macrophyte identification keys and species inventories of the area (Kylin 1949; Rueness 1977; Pankow 1990; Tolstoy et al. 2003; UoG 2010; Køie and Kristiansen 2018; Nielsen and Lundsteen 2019; Dyntaxa 2022), species that showed a potential overlap with Ulva sp. 2 were identified to assess the clades taxonomic affiliation. Individuals of Ulva sp. 2 showed morphological overlaps with certain species that were predicted to be present in the area, but that could not be verified yet due to limited genetic data available. Additionally, rbcL GenBank entries identified as Ulva ralfsii (Harvey) Le Jolis clustered within the clade delimiting Ulva sp. 2 in our phylogenetic analysis (Fig. 2) and therefore this species was included in our literature research. In the following results on the comparative literature review of the overlapping and divergent characters of above listed species will be made:
Only few sequences of Ulva clathrata are available for the rbcL gene (GenBank accession numbers: AF525939, AF525940, AY255862), however no molecular investigations on the holotype were carried out. It should be mentioned that the original type material of U. clathrata from the type location Fehmarn, Germany, Baltic Sea is flagged in algaebase as missing and that a neotype from Landskrona, Sweden, Baltic Sea has been designated (Guiry and Guiry 2022). Furthermore, the available U. clathrata sequences were not detected among the top 20 matches during NCBI BLAST searches, and there are no available reference sequences of this species for the tufA gene available yet. Species keys of the area list U. clathrata to be present in the investigated area and morphological descriptions show overlaps of U. clathrata with Ulva sp. 2 (Rueness 1977). Ulva clathrata individuals of the area are described as having rectangular or polygonal cells which have a length of 14–25 µm, yet also longer ones (up to 50 µm) in the basal part have been observed, while usually enclosing (1) 3–5 pyrenoids. These micromorphological criteria widely overlap with our findings made for Ulva sp. 2. However, other authors describe round cells in U. clathrata (Cormaci et al. 2014). Further, the tubular thallus of U. clathrata was described with a width of up to 5 mm, exhibiting an irregular branching pattern or little to no branches, whereas branchlets sometimes present an apical cell (Cormaci et al. 2014). Those traits, combined with the colour scheme makes individuals identified as U. clathrata in above named studies overlapping with the morphology described for Ulva sp. 2. However, pictures and schematic drawings of individuals identified as U. clathrata were distinctively more branched (Kylin 1949; Rueness 1977) compared to the findings made on individuals of Ulva sp. 2 (Fig. 3F–I). Another significant difference between U. clathrata and individuals of Ulva sp. 2 is that the chloroplast orientation in U. clathrata is characterized as discoidal and cell-centered (Kylin 1949; Rueness 1977; Cormaci et al. 2014) whereas the chloroplast was mainly observed to have a parietal orientation and only seldomly cell-filling in Ulva sp. 2. Additionally, individuals of U. clathrata were described with a length of up to 40 cm (Cormaci et al. 2014), which is not in accordance with Ulva sp. 2 (2–25 cm).
Individuals identified as Ulva sp. 2 furthermore shared morphological traits with Ulva kylinii, which was described at the Swedish west coast by the phycologist Carl Bliding 1948 as Enteromorpha kylinii. Young specimens of Ulva kylinii were collected in early June by Bliding (1949) outside Kristineberg research station, on the Swedish west coast where it grew on mussel shells and stones, sometimes together with by U. clathrata. Individuals of U. kylinii were described as having branches ranging from 0.5 to 3 mm in width which agrees with observations made for Ulva sp. 2. Similar to Ulva sp. 2 the rectangular or square shaped cells of U. kylinii contained two or more pyrenoids and were ordered in rows and the chloroplast orientation was described to be filling the cell or parietal which is similar with that of Ulva sp. 2 (Bliding 1949). Nevertheless, cell sizes of U. kylinii were smaller with square measurements of 16 × 16 µm and rectangular of 16 × 14 µm. Furthermore, distinctive differences among U. kylinii and Ulva sp. 2 were made in the length which can be up to 1 m in U. kylinii (Bliding 1949) and individuals of Ulva sp. 2 had a maximum length of 25 cm. Further, Bliding (1949) described that branching was only observed in the very basal parts of the thalli, which is a striking difference in comparison to Ulva sp. 2 which shows branching throughout the thallus. Comparisons of individuals identified as Ulva sp. 2 of this study and Blidings holotype of U. kylinii (LD 1,168,816) confirmed the non-matching long and unbranched thalli. However, the lectotype was too timeworn to define other traits. Currently, there is no sequenced reference material on U. kylinii and destructive sampling of Bliding´s holotype of U. kylinii (LD 1,168,816) was not possible.
An Ulva species that is not appearing in any of the investigated species keys and inventory lists of the area is Ulva ralfsii which was described from Bangor, North Wales, UK (see also Hayden et al. (2003) and Guiry and Guiry (2022)). However, rbcL reference sequences from GenBank which were identified as U. ralfsii clustered within the clade delimiting Ulva sp. 2. As is the case with U. kylinii, no molecular investigations of type material have been carried out until now and probably a neotypification is necessary for this species, as the type material is noted to be absent (Guiry and Guiry 2022). Due to the salience in the phylogenetic analysis and since U. ralfsii was recorded along the German and Dutch coasts (Guiry and Guiry 2022) we here compare the morphology of U. ralfsii with that of individuals of Ulva sp. 2.
The original description of U. ralfsii (Le Jolis 1863) described its cells as “large emerald-green granules”, which are characterized as huge and hyaline enclosing, which probably refers to thickened cell walls. More recent findings describe the cells of U. ralfsii to contain 2–6 (-8) pyrenoids (Cormaci et al. 2014) and individuals show “practically no proliferations” (Bliding 1963) and are therefore mainly unbranched which morphologically distinguishes U. ralfsii from Ulva sp. 2. Further, individuals of U. ralfsii have been observed being up to 50 cm long (Cormaci et al. 2014) which contrasts with the distinctively smaller individuals of Ulva sp. 2 (Fig. 3F–H). Notably, Bliding (1963) described that reproductive U. ralfsii tissue only released zoospores with 4 flagella and that the descendants similarly produced the same kind of swarmers as the mother generation. Our ontogenetic investigations, however, show that gametophytes of Ulva sp. 2 released biflagellate gametes (Fig. 4).
The sequence divergence of the clade representing Ulva sp. 2 from other species within the genus Ulva, in combination with the displayed morphological and ontogenetic differences, indicates that Ulva sp. 2 is genetically and morphologically distinct from other previously described species. We here describe this new species as Ulva capillata sp. nov.:
Ulva capillata S.Steinhagen, sp. nov. – HOLOTYPE: SWEDEN, Båleröd, Västra Götalands Län, N 58.890662° E 11.199227°, 25 Jun 2020, coll. S. Steinhagen (GB-0209612).
Species description
Thalli tubular, light to dark green, compressed or inflated, bearing uni- or multiseriate branches across the whole thallus (rarely unbranched), branches and microscopic appendices sharp- or blunt-ended, attached by rhizoids to substratum or free floating in shallow bays, multiple individuals originating from the same rhizoidal zone forming dense bushy tufts or single individuals, 20–250 mm (mean ± 48 mm; rarely > 20 mm) long, 0.1–2.7 mm (mean ± 0.3 mm; rarely > 2.7 mm) broad.
Cells in surface view in clear longitudinal rows throughout the whole thallus, cells square, rectangular, or polygonal with sharp angular, seldomly rounded corners, most commonly 9.3–26.4 (45) µm long and 4.8–20 µm broad. The chloroplast is mostly parietal or cell-filling and frequently located at the same side of cells within single rows, cells containing 2–5 (rarely 1 or 6) randomly located pyrenoids. Reproduction of gametophytes by biflagellate gametes. From the settled gamete a germination-tube arises which later forms the rhizoidal zone. By mitotic cell divisions a monostromatic filament develops and subsequently an erect tube is formed.
Etymology
The species name capillata refers to the hairy and bushy morphology of the species. The species name means ´hair´ or ´single hair filament´ in Latin.
GENBANK ACCESSION: OL421407 represents the sequence of the tufA marker gene and OP265117 is the respective rbcL sequence.
Type locality
Båleröd, Västra Götalands Län, Sweden (N 58.890662° E 11.199227°). Thalli were either growing as dense entangled turf in the upper littoral zone on stone or epiphytic on Fucus spp. or were free floating as entangled patches (Fig. 3A).
Other selected specimens examined (paratypes)
Hundested, Sjælland, Denmark (N 55.994062° E 11.907516°), 15 July 2020, S. Steinhagen, GenBank OP267648 (tufA) and OP265104 (rbcL), sample ID DK_020, Kattegat; Helsingør, Sjælland, Denmark (N 56.04203° E 12.61178°), 15 July 2020, S. Steinhagen, GenBank OL421114 (tufA) and OP265105 (rbcL), sample ID DK_029, Kattegat; Grevinge, Sjælland, Denmark (N 55.780297° E 11.60983°), 17 July 2020, S. Steinhagen, GenBank OL421175 (tufA) and OP265106 (rbcL), sample ID DK_096, Lammefjord; Fredrikshavn, Nordjylland, Denmark (N 57.425405° E 10.5281633°), 21 July 2020, S. Steinhagen, GenBank OL421192 (tufA) and OP265107 (rbcL), sample ID DK_113, Skagerrak/Kattegat; Fredericia, Syddanmark, Denmark (N 55.553609° E 9.727395°), 23 July 2020, S. Steinhagen, GenBank OL421233 (tufA) and OP265108 (rbcL), sample ID DK_157, Kattegat; Fredericia, Syddanmark, Denmark (N 55.553609° E 9.727395°), 23 July 2020, S. Steinhagen, GenBank OL421234 (tufA) and OP265109 (rbcL), sample ID DK_158, Kattegat; Fredericia, Syddanmark, Denmark (N 55.553609° E 9.727395°), 23 July 2020, S. Steinhagen, GenBank OP267654 (tufA) and OP265110 (rbcL), sample ID DK_159, Kattegat; Aarhus, Jütland, Denmark (N 56.138782° E 10.212697°), 23 July 2020, S. Steinhagen, GenBank OL421248 (tufA) and OP265111 (rbcL), sample ID DK_176, Kattegat; Aarhus, Jütland, Denmark (N 56.138782° E 10.212697°), 23 July 2020, S. Steinhagen, GenBank OL421249 (tufA) and OP265112 (rbcL), sample ID DK_177, Kattegat; Sønderborg, Syddanmark, Denmark (N 54.900669° E 9.794234°), 24 July 2020, S. Steinhagen, GenBank OL421282 (tufA) and OP265113 (rbcL), sample ID DK_214, Flensburg Fjord; Bagenkop, Langeland, Denmark (N 54.75111° E 10.673434°), 25 July 2020, S. Steinhagen, GenBank OL421298 (tufA) and OP265114 (rbcL), sample ID DK_232, Great Belt; Nevlunghavn, Vestfold County, Norway (N 58.967687° E 9.868426°), 3 July 2020, S. Steinhagen, GenBank OL421353 (tufA) and OP265115 (rbcL), sample ID NO_084, Skagerrak; Nevlunghavn, Vestfold County, Norway (N 58.967687° E 9.868426°), 3 July 2020, S. Steinhagen, GenBank OL421354 (tufA) and OP265116 (rbcL), sample ID NO_085, Skagerrak; Resö, Vestra Götalands Län, Sweden (N 58.7999° E 11.1654°), 18 August 2018, S. Steinhagen, GenBank OP267735 (tufA), sample ID SV_14.1, Skagerrak; Höganäs, Skåne, Sweden (N 56.198416° E 12.548323°), 5 July 2020, S. Steinhagen, GenBank OP267934 (tufA), sample ID SV_451, Kattegat; Båleröd, Vestra Götalands Län, Sweden (N 58.8917° E 11.2005°), 25 June 2020, S. Steinhagen, GenBank OP267682 (tufA) and OP265118 (rbcL), sample ID SV_760, Skagerrak; Resö, Vestra Götalands Län, Sweden (N 58.7999° E 11.1654°), 25 June 2020, S. Steinhagen, GenBank OL421416 (tufA) and OP265119 (rbcL), sample ID SV_772, Skagerrak; Sannäs, Vestra Götalands Län, Sweden (N 58.7417° E 11.2456°), 25 June 2020, S. Steinhagen, GenBank OL421421 (tufA) and OP265120 (rbcL), sample ID SV_777, Skagerrak; Marstrand, Vestra Götalands Län, Sweden (N 57.887513° E 11.587307°), 30 July 2020, S. Steinhagen, GenBank OL421450 (tufA) and OP265121 (rbcL), sample ID SV_808, Skagerrak.
Discussion
Our results demonstrate that due to the crypticity of tubular entities of the genus Ulva, present in the Atlantic-Baltic Sea transect, certain species remained undetected until recently. One of these entities is here described as Ulva capillata sp. nov. which appears to be a relatively frequent species in the Atlantic-Baltic Sea transect.
We support the description of U. capillata as a new species within the genus Ulva with three main lines of evidence including (1) molecular evidence and clear delimitation of individuals of U. capillata from closely related species, (2) morphological delimitation by a distinctive habitus and incisive identification criteria from species expected to be present in the area (e.g. U. clathrata, U. kylinii, U. ralfsii), and (3) significant differences in ontogenetic developmental patterns with potentially conspecific species (e.g. U. ralfsii). Our results strongly emphasize the importance and necessity to molecularly investigate especially tubular entities and their respective historic type vouchers within the genus Ulva. A clear species identification is often hindered by the absence of molecular data of holo- and lectotype specimens, as well as sometimes vague type descriptions and missing or unavailable type material for observation. Such missing vital data adds up to the taxonomic confusion within the genus Ulva and supports the ongoing mis-application of names of e.g. cryptic species concepts and therefore fosters the continuous changes in the systematics of this green algae group.
During an extensive field survey on the molecular biodiversity of Ulva spp. in the Atlantic-Baltic Sea transect (see also Steinhagen et al. 2022c and Steinhagen et al. 2019a) a revised picture of the diversity and distribution of the genus Ulva in the respective area was carried out and species-specific range margins were defined. It became obvious that several species, species-complexes, and singletons could not be identified to species level, due to the absence of reference material, whereas other clades contained sequences with name applications of different species concepts and therefor reflected polyphyletic species groups (Steinhagen et al. 2019a, 2022c). Such difficulties of name applications have been recently discussed for several of the most commonly applied molecular markers that are used in Ulva identification and delimitation (Tran et al. 2022). One large problem during molecular identification is the inconsistency of species name applications to sequences uploaded to genetic repositories such as GenBank and the reality that often sequence names are not updated in said databases once the taxonomy or systematics of respective species have changed. Furthermore, a central and utmost important point for correct identification within genera that maintain several cryptic species, as is the case within Ulva (e.g. Brodie et al. 2003; Steinhagen et al. 2019a, b; Fort et al. 2021; Hughey et al. 2022), is the necessity of the availability of molecularly investigated type matrial. Such genetic information of lecto-, holo-, or neotype vouchers however, is very limited within the genus Ulva. Whereas recent studies made important and valuable contributions of molecular data generated on type material of mainly leaf-like Ulva species (Steinhagen et al. 2019a; Fort et al. 2021, 2022; Hughey et al. 2022; Tran et al. 2022) such vital information is mainly absent for most tubular Ulva species. Therefore, molecular information on tubular Ulva spp. is mostly limited to recently described species (e.g. Chen et al. 2015; Lagourgue et al. 2022). The gap of missing data on type material becomes especially obvious when looking on the many homotypic and heterotypic synonyms that have been listed for the 85 taxonomically valid Ulva species (Guiry and Guiry 2022). However, generating genetic information of type specimens will be unfeasible for lost or degraded vouchers or specimens with unknown location (Tran et al. 2022). Therefore, it needs to be discussed, if a tabula rasa approach which facilitates the designation of epi- or neotypes or the description of new species could be a suitable solution (see also Tran et al. 2022).
In describing a new tubular Ulva species here, namely U. capillata, we provide detailed molecular and morphological information for an entity which can frequently be found in fully marine conditions of the Atlantic as well as brackish water conditions of the SW Baltic Sea and which was potentially widely confused with morphologically overlapping species of the area. Our data suggest that this discrepancy results from the exclusive use of morphological traits and ontogenetic developmental characters as identification criteria in the past. During phylogenetic analyses our study revealed that specimens identified as U. capillata resolved in well delimited clades, for both, the rbcL and tufA genes. While there was no reference sequence clustering for the cluster delimiting U. capillata within the tufA phylogeny, such uniqueness of sequence was not observed for the rbcL gene, and individuals form New Zealand identified as U. ralfsii were nearly identical with sequences from U. capillata. The original genetic data on specimens identified as U. ralfsii, originataing from New Zealand, did not include detailed macro-morphological or cytological characteristics and main focus was on valuable aquaculture and bioremediation purposes of different Ulva spp., not explicitly on resolving taxonomic patterns within the genus Ulva (Kidgell et al. 2021; Lawton et al. 2021).
Therefore, extensive morphological studies on the northern Hemisphere individuals of U. capillata were carried out, to determine its taxonomic identity and potential morphological overlaps with the species concept of U. ralfsii (Le Jolis 1863). Notably, distinct morphological differences of U. ralfsii and U. capillata were detected. Whereas U. ralfsii was described as mainly unbranched and very fine-filamentous, individuals of U. capillata were mainy branched and even though having a bushy and hairy morphology the habitus differed from U. ralfsii (Le Jolis 1863). Furthermore, the thalli of U. ralfsii were observed to be very long compared to any of the samples from the Swedish coast. Additionally, U. ralfsii has never been identified in Swedish waters before and is not listed in any of the species lists or identififcation keys of the area (Kylin 1949; Rueness 1977; Pankow 1990; Tolstoy and Österlund 2003; UoG 2010; Køie and Kristiansen 2018; Nielsen and Lundsteen 2019; Dyntaxa 2022). Even though a DNA match in overlapping sequences could be considered the most evident proof of conspecificity, it is most important to have reliable reference material, preferably of lecto-, holo-, or neotype vouchers. Unfortunately, such molecular information of type material is absent for U. ralfsii. However, the discrepancies in morphological characters makes a conspecificity of U. ralfsii and U. capillata unlikely. It should however be noted that U. capillata probably has a globally ubiquitous distribution, since three individuals with similar rbcL sequence reads have been recorded from New Zealand.
Since there was a strong discrepancy discovered among historical species inventories and molecularly validated Ulva species of the area (Steinhagen et al. 2019a, b, c, 2022c) morphological data obtained on U. capillata within this study was also compared to listed species of the area which have not been identified molecularly, including those species which lack any genetic reference data. Especially U. clathrata and U. kylinii which were recorded to occur in the Atlantic-Baltic Sea transect, (Bliding 1948; UoG 2010; Nielsen and Lundsteen 2019; Dyntaxa 2022), were identified to share certain traits with U. capillata. Morphological investigations however revealed distinct differences and we therefore excluded conspecificity with any of the species. It should however be noted, that for both U. clathrata and U. kylinii no molecular data of the type vouchers were available.
We can conclude that the historical species concepts for Ulva spp. are still flawed and problematic, especially for tubular species. Misinterpretation due to phenotypic plasticity has led to misidentifications in the past, and species delimitation based on morphological traits is often impossible as shown in this study. Thus, our findings support not only the use of molecular methods for correct and clear species identification and deemphasize the use of morphological characters alone, but also highlights once more that molecular investigations of type material within the genus Ulva is urgently required to stop the continuing changes of the taxonomy within this ubiquituous as well as ecologically and economically important green algae genus.
Data availability
DNA sequences of the tufA and rbcL gene of the examined specimens are available from GenBank (accession numbers: see Table 1). All other data generated or analysed during this study are included in this published article.
References
Bliding C (1949) Enteromorpha kylini, eine neue Art aus der schwedischen Westküste. Kungl Fisiografiska Sallsk Lund Forhandl 18:199–204
Bliding C (1963) A critical survey of European taxa in Ulvales. Part 1. Capsosiphon, Percursaria, Blidingia, Enteromorpha. Opera Bot 8:1–160
Blomster J, Bäck S, Fewer DP, Kiirikki M, Lehvo A, Maggs CA, Stanhope MJ (2002) Novel morphology in Enteromorpha (Ulvophyceae) forming green tides. Am J Bot 89:1756–1763
Blomster J, Maggs CA, Stanhope MJ (1998) Molecular and morphological analysis of Enteromorpha intestinalis and E. compressa (Chlorophyta) in the British Isles. J Phycol 34:319–340
Bolton JJ, Robertson-Andersson DV, Shuuluka D, Kandjengo L (2009) Growing Ulva (Chlorophyta) in integrated systems as a commercial crop for abalone feed in South Africa: a SWOT analysis. J Appl Phycol 21:575–583
Bolton JJ, Cyrus MD, Brand MJ, Joubert M, Macey BM (2016) Why grow Ulva? Its potential role in the future of aquaculture. Perspect Phycol 3:113–120
Brodie J, Maggs CA, John DM (eds) (2007) The green seaweeds of Britain and Ireland. The British Phycological Society, UK
Cai C, Gu K, Zhao H, Steinhagen S, He P, Wichard T (2021) Screening and verification of extranuclear genetic markers in green tide algae from the Yellow Sea. PloS ONE 16:e0250968
Califano G, Kwantes M, Abreu MH, Costa R, Wichard T (2020) Cultivating the macroalgal holobiont: effects of integrated multi-trophic aquaculture on the microbiome of Ulva rigida (Chlorophyta). Front Mar Sci 7:52
Carl C, de Nys R, Lawton RJ, Paul NA (2014) Methods for the induction of reproduction in a tropical species of filamentous Ulva. PLoS ONE 9:e97396
Chen L, Feng J, Xie SL (2015) Ulva shanxiensis (Ulvaceae), a new species from Shanxi, China. Novon: J Bot Nomenclature 23:397–405
Cormaci M, Furnari G, Alongi G (2014) Flora marina bentonica del Mediterraneo: Chlorophyta. Bull Gioenia Acad Nat Sci Catania 47:FP11–FP436
Dyntaxa (2022) Swedish taxonomic database. www.dyntaxa.se. Accessed 29 June 2022
Fort A, McHale M, Cascella K, Potin P, Usadel B, Guiry MD, Sulpice R (2021) Foliose Ulva species show considerable inter-specific genetic diversity, low intra-specific genetic variation, and the rare occurrence of inter-specific hybrids in the wild. J Phycol 57:219–233
Fort A, McHale M, Cascella K, Potin P, Perrineau M-M, Kerrison PD, da Costa E, Calado R, Domingues MDR, Costa Azevedo I, Sousa-Pinto I, Gachon C, van der Werf A, de Visser W, Beniers JE, Jansen H, Guiry MD, Sulpice R (2022) Exhaustive reanalysis of barcode sequences from public repositories highlights ongoing misidentifications and impacts taxa diversity and distribution. Mol Ecol Resour 22:86–101
Guiry MD, Guiry GM (2022) AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. https://www.algaebase.org; searched on 12. February 2022
Hayden HS, Blomster J, Maggs CA, Silva PC, Stanhope MJ, Waaland JR (2003) Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. Eur J Phycol 38:277–294
Hoeksema BW, Van den Hoek C (1983) The taxonomy of Ulva (Chlorophyceae) from the coastal region of Roscoff (Brittany, France). Bot Mar 26:65–86
Hughey JR, Maggs CA, Mineur F, Jarvis C, Miller KA, Shabaka SH, Gabrielson PW (2019) Genetic analysis of the Linnaean Ulva lactuca (Ulvales, Chlorophyta) holotype and related type specimens reveals name misapplications, unexpected origins, and new synonymies. J Phycol 55:503–508
Hughey JR, Gabrielson PW, Maggs CA, Mineur F (2022) Genomic analysis of the lectotype specimens of European Ulva rigida and Ulva lacinulata (Ulvaceae, Chlorophyta) reveals the ongoing misapplication of names. Eur J Phycol 57:143–153
Le Jolis A (1863) Liste des algues marines de Cherbourg. Mém Soc Imp Sci Nat Cherbourg 10:5–168, pls I-IV
Katoh K, Misawa K, Miyata K-I, T, (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl Acids Res 30:3059–3066
Kidgell JT, Carnachan SM, Magnusson M, Lawton RJ, Sims IM, Hinkley SF, de Mys R, Glasson CR (2021) Are all ulvans equal? A comparative assessment of the chemical and gelling properties of ulvan from blade and filamentous Ulva. Carbohydr Polym 264:118010
Koeman RT, Van Den Hoek C (1984) The taxonomy of Enteromorpha Link, 1820 (Chlorophyceae) in the Netherlands. III. The sections Flexuosae and Clathratae and an addition to the section Proliferae. Cryptogam Algol 5:21–61
Koeman RPT, van den Hoek C (1981) The taxonomy of Ulva (Chlorophyceae) in the Netherlands. Brit Phycol J 16:9–53
Koeman RPT, Van den Hoek C (1982a) The taxonomy of Enteromorpha Link, 1820, (Chlorophyceae) in the Netherlands. I: The section Enteromorpha. Algol Stud 32:279–330
Koeman RPT, Van Den Hoek C (1982b) The taxonomy of Enteromorpha Link, 1820, (Chlorophyceae) in the Netherlands. II: the section Proliferae. Cryptogam Algol 3:37–70
Køie M, Kristiansen A (2018) Havets djur och växter. Gyldendal, Copenhagen
Kraft LGK, Kraft GT, Waller RF (2010) Investigations into southern Australian Ulva (Ulvophyceae, Chlorophyta) taxonomy and molecular phylogeny indicate both cosmopolitanism and endemic cryptic species. J Phycol 46:1257–1277
Krupnik N, Paz G, Douek J, Lewinsohn E, Israel A, Carmel N, Mineur F, Maggs CA (2018) Native, invasive and cryptogenic Ulva species from the Israeli Mediterranean Sea: risk and potential. Med Mar Sci 19:132–146
Kylin H (1949) Die Chlorophyceen der schwedischen Westküste. Lunds Univ Arsskr NF Avd 2 45(4):1–79
Lagourgue L, Gobin S, Brisset M, Vandenberghe S, Bonneville C, Jauffrais T, Van Wynsberge S, Payri CE (2022) Ten new species of Ulva (Ulvophyceae, Chlorophyta) discovered in New Caledonia: genetic and morphological diversity, and bloom potential. Eur J Phycol 57:458–478
Lawton RJ, Sutherland JE, Glasson CR, Magnusson ME (2021) Selection of temperate Ulva species and cultivars for land-based cultivation and biomass applications. Algal Res 56:102320
Linnaeus C (1753) Species Plantarum, Exhibentes Plantas Rite Cognitas Ad Genera Relatas: Cum Differentiis Specificis, Nominibus Trivialibus, Synonymis Selectis, Locis Natalibus, Secundum Systema Sexuale Digestas. Trattner, Stockholm
Nielsen R, Lundsteen S (2019) Danmarks Havalger Bind 3 Grønalger Chlorophyta). Scientia Danica. Series B, Biologica 7. Det Kongelige Danske Videnskabernes Selskab, København
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University
Olsson J, Raikova S, Mayers JJ, Steinhagen S, Chuck CJ, Nylund GM, Albers E (2020) Effects of geographical location on potentially valuable components in Ulva intestinalis sampled along the Swedish coast. Appl Phycol 1:80–92
Pankow H (1990) Ostsee-Algenflora. G. Fischer Verlag, Jena
Phillips JA, Lawton RJ, Denys R, Paul NA, Carl C (2016) Ulva sapora sp. nov., an abundant tubular species of Ulva (Ulvales) from the tropical Pacific Ocean. Phycologia 55:55–64
Provasoli L (1968) Media and prospects for the cultivation of marine algae. In: Cultures and Collections of Algae. Proceedings of the 1968 US-Japan Conference Society of Plant Physiology, Hakone, pp 12–15
Rosenberg G, Ramus J (1984) Uptake of inorganic nitrogen and seaweed surface area: volume ratios. Aquat Bot 19:65–72
Rueness J (1977) Norsk algeflora. Universitetsforl.
Shimada S, Hiraoka M, Nabata S, Iima M, Masuda M (2003) Molecular phylogenetic analyses of the Japanese Ulva and Enteromorpha (Ulvales, Ulvophyceae), with special reference to the free-floating Ulva. Phycol Res 51:99–108
Smetacek V, Zingone A (2013) Green and golden seaweed tides on the rise. Nature 504:84–88
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Steinhagen S, Karez R, Weinberger F (2019a) Cryptic, alien and lost species: molecular diversity of Ulva sensu lato along the German coasts of the North and Baltic Seas. Eur J Phycol 54:466–483
Steinhagen S, Weinberger F, Karez R (2019b) Molecular analysis of Ulva compressa (Chlorophyta, Ulvales) reveals its morphological plasticity, distribution and potential invasiveness on German North Sea and Baltic Sea coasts. Eur J Phycol 54:102–114
Steinhagen S, Enge S, Larsson K, Olsson J, Nylund GM, Albers E, Pavia H, Undeland I, Toth GB (2021) Sustainable large-scale aquaculture of the northern hemisphere sea lettuce, Ulva fenestrata, in an off-shore seafarm. J Mar Sci Eng 9:615
Steinhagen S, Larsson K, Olsson J, Albers E, Undeland I, Pavia H, Toth GB (2022b) Closed life-cycle aquaculture of sea lettuce (Ulva fenestrata): performance and biochemical profile differ in early developmental stages. Front Mar Sci 9:942679
Steinhagen S, Karez R, Weinberger F (2019c) Surveying seaweeds from the Ulvales and Fucales in the world’s most frequently used artificial waterway, the Kiel Canal. Bot Mar 62:51–61
Steinhagen S, Enge S, Cervin G, Larsson K, Edlund U, Schmidt AE, Wahlström N, Kollander B, Pavia H, Undeland I, Toth GB (2022a) Harvest time can affect the optimal yield and quality of sea lettuce (Ulva fenestrata) in a sustainable sea-based cultivation. Front Mar Sci 9:816890
Steinhagen S, Hoffmann, S, Pavia H and Toth GB (2022c) Large scale biodiversity monitoring of the genus Ulva in the Atlantic-Baltic Sea gradient. Under review at PNAS
Teichberg M, Fox SE, Olsen YS, Valiela I, Martinetto P, Iribarne O, Muto EY, Petti MA, Corbisier TN, Soto-Jiménez M, Pàez-Osuna F, Castro P, Freitas H, Zitteli A, Cardinaletti M, Tagliapietra D (2010) Eutrophication and macroalgal blooms in temperate and tropical coastal waters: nutrient enrichment experi- ments with Ulva spp. Global Change Biol 16:2624–2637
Tolstoy A, Österlund K, Ankar S, Persson M (2003) Alger vid Sveriges Östersjökust: en fotoflora. ArtDatabanken, Uppsala
Tran L-AT, Vieira C, Steinhagen S, Maggs CA, Hiraoka M, Shimada S, Van Nguyen T, De Clerck O, Leliaert F (2022) An appraisal of Ulva (Ulvophyceae, Chlorophyta) species diversity, species delimitation, and taxonomy. J Appl Phycol 34:2689–2703
UoG (2010) Identification key of macrophytes of the University of Gothenburg.
Acknowledgements
We would like to express our thanks to the Herbarium of the University of Gothenburg for their close cooperation and lodging the vouchers of this study. Furthermore, we thank Samanta Hoffman for her intense support during field collections.
Funding
Open access funding provided by University of Gothenburg. No specific funding to declare. The authors thank the Formas-funded ‘A manual for the use of sustainable marine resources’ project (Grant no. 2022-00331) for financial support.
Author information
Authors and Affiliations
Contributions
S. Steinhagen: original concept, experimental design, fieldwork and algae collection, laboratory work, macro- and microscopic observation, phylogenetic analysis, drafting and editing manuscript, supervision degree project L. Kramar; L. Kramar: algae collection, laboratory work, morphological investigations and algae cultivation; G. Toth: algae collection, funding acquisition, editing of manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Steinhagen, S., Kramár, L. & Toth, G.B. The unheeded existence of the tubular greens: molecular analyses reveal the distribution of a new Ulva species (Ulvophyceae, Chlorophyta), Ulva capillata sp. nov. in the Atlantic-Baltic Sea transect. J Appl Phycol 35, 509–522 (2023). https://doi.org/10.1007/s10811-022-02886-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-022-02886-w