Resurrection of the Family Grateloupiaceae Emend. (Halymeniales, Rhodophyta) Based on a Multigene Phylogeny and Comparative Reproductive Morphology
- 1Research Institute of Basic Sciences, Jeju National University, Jeju, South Korea
- 2Korea Inter-University Institute of Ocean Science, Pukyong National University, Busan, South Korea
- 3Marine Ecosystem Research Center, Korea Institute of Ocean Science and Technology, Busan, South Korea
- 4Department of Biology, Chungnam National University, Daejeon, South Korea
- 5Department of Biological Sciences, University of Alabama, Tuscaloosa, AL, United States
- 6Department of Biology, University of Louisiana, Lafayette, LA, United States
- 7National Institute of Water and Atmospheric Research, Auckland, New Zealand
- 8Department of Biological Sciences, Sungkyunkwan University, Suwon, South Korea
- 9Department of Biology, Jeju National University, Jeju, South Korea
The marine red algal order Halymeniales currently includes two families, the Halymeniaceae and Tsengiaceae, and consist of 38 genera and about 358 species. Phylogenetic analyses on specific taxa of the order are common, but not comprehensive, leaving the many intra-ordinal relationships within the Halymeniales unresolved. To reassess the phylogeny of the Halymeniales, we conducted extensive phylogenetic analyses based on 207 rbcL sequences and multigene analyses (rbcL, psaA, psbA, cox1, and LSU) using 47 taxa from the order. The combined data set fully supports the monophyly of the Grateloupia sensu lato clade. Phylogenetic assessment of the reproductive structures in the order using the type of auxiliary cell ampullae, pericarp origin, and tetrasporangial development characters, supports a Grateloupia sensu lato clade distinct from the Halymeniaceae exemplified by the generitype Halymenia. As a result, we propose to reinstate the family Grateloupiaceae Schmitz based on the Grateloupia sensu lato clade and including Grateloupia and eight other genera: Dermocorynus, Mariaramirezia, Neorubra, Pachymeniopsis, Kintokiocolax, Phyllymenia, Prionitis, and Yonagunia. The emended Grateloupiaceae is distinguished from the Halymeniaceae by the following three characteristics; (i) simple unbranched and unilateral type of auxiliary cell ampullae, (ii) pericarp formed densely by the fusion of secondary medullary filaments from subcortical cells and lateral ampullary filaments from a fusion cell complex, (iii) tetrasporangia originating laterally from the outer cortex. The Halymeniales comprises the monophyletic Grateloupiaceae, Halymeniaceae sensu lato (which requires further study), and the Tsengiaceae.
Introduction
The monophyletic Rhodophyta has been divided into seven classes based on multigene data (Yoon et al., 2006). Among the seven classes, the Florideophyceae is the largest and well known red algal class, comprising 95% of currently described species (Guiry and Guiry, 2021). A series of molecular phylogenetic studies by Saunders et al. (2002), Withall and Saunders (2006), and Le Gall and Saunders (2007) provided many improvements for resolving relationships among florideophyte orders. Recently, the close relationships of Halymeniales, Sebdeniales, and Rhodymeniales were confidently resolved based on multigene and mitochondrial genome data (e.g., Yang et al., 2015, 2016). However, to date, there have been no extensive molecular phylogenetic studies of the Halymeniales as a whole.
In 1996 Saunders and Kraft recognized a new name, the Halymeniales, for the red algal order that was previously known as the Cryptonemiales Kylin, based on small-subunit rRNA phylogenetic analyses. A historical account of the modified taxonomic concepts with regard to the Cryptonemiales was provided in detail by Silva and Johansen (1986), Silva (2002), and Krayesky et al. (2009). The emended order Halymeniales first accommodated the families Halymeniaceae Bory and Sebdeniaceae Kylin, and was characterized by taxa with a multiaxial thallus, non-procarpic female reproductive development, outwardly directed carpogonial branches and intercalary auxiliary cells (Saunders and Kraft, 1996). Subsequently, Saunders and Kraft (2002) transferred the genus Tsengia K. C. Fan and Y. C. Fan from the Nemastomatales to the Halymeniales with the establishment of the new monogeneric family Tsengiaceae. The Sebdeniaceae was later removed from the Halymeniales and elevated to a new order, Sebdeniales Withall and Saunders (Withall and Saunders, 2006) as suggested by Gavio et al. (2005). In the current molecular taxonomic framework, the order Halymeniales includes two families, the Halymeniaceae and Tsengiaceae, consisting of 38 genera and about 358 species (Guiry and Guiry, 2021).
The Halymeniaceae, which includes most of the taxa in the Halymeniales except for 12 species of Tsengiaceae, was established based on the genus Halymenia C. Agardh along with four other genera (Bory de Saint-Vincent, 1828). Harvey (1849) suggested the family Cryptonemiaceae based on the tribe Cryptonemieae J. Agardh, including Cryptonemia J. Agardh, Grateloupia C. Agardh, Halymenia, and 21 other genera. The family name Grateloupiaceae was proposed by Schmitz (1889), as a part of the suborder Cryptoneminae, based on 11 genera (Grateloupia, Cryptonemia, Halymenia, Aeodes J. Agardh, Carpopeltis F. Schmitz, Corynomorpha J. Agardh, Dermocorynus P. Crouan and H. Crouan, Pachymenia J. Agardh, Polyopes J. Agardh, Prionitis J. Agardh, and Thamnoclonium Kützing). Later, Papenfuss (1955) synonymized the Grateloupiaceae with the Cryptonemiaceae; however, Guiry (1978) reinstated the Halymeniaceae Bory based on nomenclatural priority. As a consequence, the Grateloupiaceae and Cryptonemiaceae have been considered synonyms of the Halymeniaceae.
A diagnostic feature of the Halymeniaceae is the presence of auxiliary cell ampullae and the special accessory branch system in the female reproductive structure (Chiang, 1970). The details of the female reproductive morphology, such as the type and development of the auxiliary cell ampullae before or after fertilization, have been regarded as a delimiting character among halymeniacean genera (Kawaguchi et al., 2004; De Clerck et al., 2005a; Lin et al., 2008; Gargiulo et al., 2013). The other female reproductive features, such as the absence or presence of a pericarp, location of carpogonial branches, and number of cells in carpogonial branch ampullae, have also been suggested as potentially diagnostic for genera in the Halymeniaceae (e.g., Balakrishnan, 1961a; Hommersand and Fredericq, 1990; Kawaguchi et al., 2004; Rodríguez-Prieto et al., 2018). In addition, monoecy or dioecy of the gametophyte was referred to as one of the noticeable characteristics for inferring the phylogenetic relationships within the Halymeniaceae (Guiry and Maggs, 1982; Womersley and Lewis, 1994; Lee and Kim, 2019). In contrast, vegetative features such as overall habit and thallus texture show little taxonomic utility in the family (Wang et al., 2001; Wilkes et al., 2005; Rodríguez-Prieto et al., 2018).
The genus Grateloupia C. Agardh is the largest and most taxonomically complex group in the Halymeniaceae (Gargiulo et al., 2013). In the early 2000s, several genera were merged into Grateloupia based on rbcL phylogenetic analyses and Grateloupia-type auxiliary cell ampullae (Wang et al., 2001; De Clerck et al., 2005b; Wilkes et al., 2005). However, Lin et al. (2008) suggested two differentiating types of auxiliary cell ampullae (Grateloupia taiwanensis- and G. orientalis- type) and emphasized the importance of careful observation of the developmental stages. Subsequently, Gargiulo et al. (2013) provided a new interpretation of this clade in terms of the type of ampullae and post-fertilization events, suggesting that some species be separated into distinct genera. As a result, the genera, i.e., Dermocorynus, Prionitis, Pachymeniopsis Yamada ex Kawabata, and Phyllymenia J. Agardh, which had been subsumed into Grateloupia by previous authors, were resurrected. Despite the resurrection of several genera, many species still have the generic name “Grateloupia.” In the historical context of this classification, Grateloupia sensu lato includes not only Grateloupia species, but also Dermocorynus, Prionitis, Pachymeniopsis, and Phyllymenia. In addition, Kintokiocolax Tak.Tanaka and Y. Nozawa and Yonagunia S. Kawaguchi and M. Masuda have been regarded as members of Grateloupia sensu lato based on the rbcL phylogenetic analyses (Calderon et al., 2014a; Yang and Kim, 2015). Later, two new genera, Neorubra M. S. Calderon, G. H. Boo, and S. M. Boo and Mariaramirezia M. S. Calderon, G. H. Boo, A. Mansilla, and S. M. Boo, were added to the Grateloupia sensu lato clade (Calderon et al., 2014a, 2016).
Regarding earlier systematic studies, two critical points have yet to be resolved in the Halymeniales. First, molecular studies were conducted primarily using only plastid rbcL gene sequences, a gene that has been shown to be promising for the taxonomic study of Halymeniales (e.g., Lin et al., 2008; Gargiulo et al., 2013; Calderon et al., 2014a, b). Multigene phylogenies are needed to examine deep nodes within the order (i.e., resolving family-level taxonomy), as shown in other groups of red algae (e.g., Dixon et al., 2015; Boo et al., 2016; Entwisle et al., 2018). Second, most studies have focused on few species and genera, so taxon sampling is limited. Further studies involving more species from a suite of genera are required in order to increase our understanding on the phylogeny of the Halymeniales.
In this study, we reconstructed the phylogenies of the Halymeniales by using (i) a taxon-rich rbcL data set with 45 new sequences and (ii) representative taxa using five genes (plastid rbcL, psaA, psbA, mitochondrial cox1, and nuclear LSU rRNA) with 153 new sequences for a combined data set analysis to test the monophyly of the Grateloupia sensu lato clade and to clarify the phylogenetic relationships among the clade and other genera of the Halymeniales. In addition (iii) morphological observations focused on the reproductive structures to clarify the taxonomic delimitation corresponding to our multigene phylogeny, especially to enable the distinction between Grateloupia sensu lato from the Halymeniaceae (exemplified by the generitype Halymenia). Consequently, herein we reinstate the family Grateloupiaceae, which was previously established by Schmitz (1889), and emend the taxonomic concept of the Grateloupiaceae to include nine genera.
Materials and Methods
Taxon Sampling and Analyses of Multigene Data Set
For the most extensive phylogenetic analysis, we collected a total of 207 rbcL sequences of the Halymeniales including 45 newly generated and 162 available sequences from NCBI1 and BOLD2 databases. To include qualified sequences, the downloaded sequences were checked one by one, and the results were presented as shown in Supplementary Table 1 including the reference paper and taxonomic note of each sequence. The five outgroup taxa (three Rhodymeniales and two Sebdeniales) were selected because they were shown to be sister taxa to the Halymeniales in previous studies (Withall and Saunders, 2006; Yang et al., 2016). The data set included all genera in the Halymeniales (i.e., 33 genera; Figure 1) except five uncertain genera (Blastophye J. Agardh, Grateloupiocolax Schnetter and Bula-Meyer, Hymenophlea J. Agardh, Neoabbottiella Perestenko, and Zanardinula De Toni—see Discussion for the details) and 30 type species. Five molecular markers were chosen from three genomes (i.e., plastid rbcL, psaA, psbA, mitochondrial cox1, and nuclear LSU rRNA) to infer a robust phylogeny of the Halymeniales.
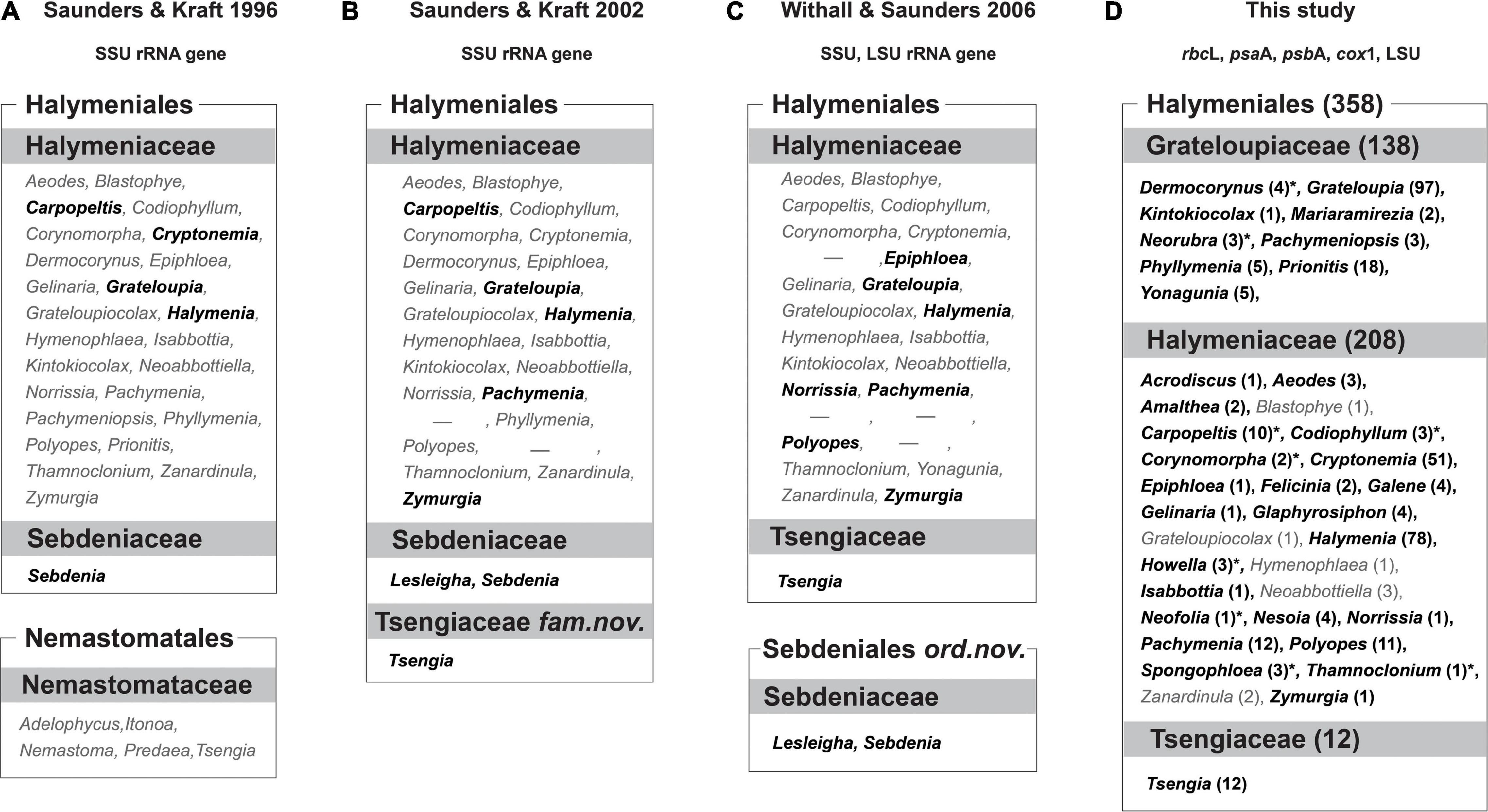
Figure 1. Comparison between current and previous taxonomic concepts of the Halymeniales. (A) Taxonomic concept of Saunders and Kraft (1996). (B) Taxonomic concept of Saunders and Kraft (2002). (C) Taxonomic concept of Withall and Saunders (2006). (D) Taxonomic concept of this study. Bolded black colored genera indicate the taxa analyzed for each study, and gray colored genera indicate the taxa not analyzed for each study. The numbers in parentheses indicate the number of species in each genus. Asterisk (*) indicates analyzed taxa for rbcL only. Dash (-) indicates the genera that have been synonymized with the genus Grateloupia but recently resurrected.
For the data-rich five-gene analysis, a total of 153 new sequences were determined. The following 42 taxa of Halymeniales were used: 24 taxa of Grateloupia sensu lato, 18 taxa of the other Halymeniaceae, and Tsengiaceae (Supplementary Table 2) representing various lineages within the order. In most cases (41 out of 47 taxa), the genes from the same specimen were used for the combined data set. The combined data set included 25 genera and 19 type species from the Halymeniales (Supplementary Table 2).
Total genomic DNA of each sample was extracted from silica gel-dried specimens using a LaboPassTM Tissue Mini kit (Hokkaido System Science, Hokkaido, Japan) following the manufacturer’s instructions. Primer pairs for amplification and sequencing of each gene were as follows; plastid rbcL- rbcLF145/rbcLR898 and rbcLF762/rbcLR1442 (Kim et al., 2010), plastid psbA- F1/R2 (Yoon et al., 2002), plastid psaA-130F/1110R (Yoon et al., 2002; Yang and Boo, 2004), mitochondrial cox1- F115Haly/R702Haly (F115Haly: 5′-ATG TCW ATG YTA ATY CGY ATG GAA-3′, R702Haly: 5′-TAA ATG TTG ATA TAA DAC WGG ATC-3′), and nuclear LSU- X/28F (Freshwater et al., 1999). PCR amplification was performed using TaKaRa Ex Taq DNA polymerase (Takara Shuzo, Shiga, Japan). PCR was carried out with an initial denaturation at 94°C for 4 min, followed by 35 cycles of amplification (denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min), with a final extension at 72°C for 6 min. The PCR products were purified with the LaboPassTM PCR kit (Hokkaido System Science, Hokkaido, Japan) and then sequenced commercially (Macrogen Inc., Seoul, South Korea). Both the forward and reverse electropherograms from each sample were edited using the program Chromas version 1.45.3
Sequences were aligned manually using Se-Al v.2.0a11 (Rambaut, 2002). A total of 207 taxa, and 1,467 bp each were used for the rbcL analyses. For the individual gene analyses and 5-gene concatenated data analyses, five individual gene alignments (rbcL: 47 taxa, 1,227 bp; psaA: 41 taxa, 927 bp; psbA: 42 taxa, 876 bp; cox1: 41 taxa, 531 bp; LSU: 44 taxa, 1,014 bp) were generated. Phylogenetic analyses were performed with the Maximum Likelihood (ML) with RAxML (Stamatakis, 2006) optimality criteria using the GTR + Γ evolutionary model. Two-hundred independent tree inferences were performed with the default option with automatically optimized SPR rearrangement and 25 distinct rate categories in the program to identify the best tree. To generate bootstrap values for the best phylogeny, 1,000 replications under the same model settings were employed. The concatenated data set was partitioned by gene using –q option. Tree topologies were examined and visualized using FigTree v.1.4.3 (Rambaut, 2009).
Morphological Observations
The reproductive morphologies of five representative species were compared within Grateloupia sensu lato and Halymenia floresii (Clemente) C. Agardh, the type species of the Halymeniaceae. The five species were chosen based on a rbcL phylogenetic analysis (Supplementary Figure 1) from three different genera as follows: Grateloupia catenata Yendo from the Yellow Sea, South Korea (MSK180814-01, MSK180814-02), a member of the genus Grateloupia sensu stricto; “Grateloupia” jejuensis S. Y. Kim, E. G. Han, and S. M. Boo from Jeju, South Korea (MSK170722-32, MSK170722-37), and “G.” livida (Harvey) Yamada from Hokkaido (MSK140224-22) and Shizuoka (MSK170417-67), Japan, members of the genus Prionitis (see Gargiulo et al., 2013); Pachymeniopsis lanceolata (K. Okamura) Y. Yamada ex S. Kawabata from Jeju, Korea (MSK150502-05, MSK161126-01), the generitype, and Pa. elliptica (Holmes) Yamada, a representative member of Pachymeniopsis from Shizuoka, Japan (MSK170417-57, MSK170417-58). The type species of the genus Grateloupia (G. filicina), studied in detail by Gargiulo et al. (2013), was also referenced and used for morphological comparison. Specimens were preserved in 2.5% Formalin/seawater for anatomical study. Sections were manually prepared using a razor blade or a bench-top freezing microtome (NK-101-II; Nippon Optical Works Co., Ltd., Tokyo, Japan). Sectioned material was stained either with 1% aniline blue acidified with 1% HCl and mounted in 35% custom-made corn syrup or stained with Wittmann’s aceto-iron- hematoxylin-chloral hydrate (Wittmann, 1965) and mounted in 50% Hoyer’s mounting medium (Hommersand and Fredericq, 1990; Lin et al., 2012). Photomicrographs were obtained using a BX43 microscope (Olympus, Tokyo, Japan) with an EOS 600D digital camera (Canon, Tokyo, Japan). Digitized images were adjusted for clarity using Adobe Photoshop software (Adobe Systems Inc., San Jose, CA, United States; ver. 6.1).
Results
Molecular Assessment
In the 207 rbcL sequence analyses, the three species of Tsengia formed a well-supported monophyletic group (98% ML bootstrap support value, MLB; Figure 2) and diverged first within the Halymeniales with 60% MLB. All the other halymenialean taxa were grouped into a second clade (81% MLB). Within the second clade, a monophyletic Grateloupia sensu lato clade was recovered with high bootstrap support (91% MLB). The support value for the Grateloupia sensu lato clade was higher than that for the other Halymeniales clade, i.e., Halymeniaceae sensu lato. The Grateloupia sensu lato clade was comprised of nine genera: Pachymeniopsis, Kintokiocolax, Prionitis, Neorubra, Phyllymenia, Grateloupia, Yonagunia, Dermocorynus, and Mariaramirezia. Among them, the monophyly of five genera was fully supported (100% MLB; clade a—Prionitis, clade b—Neorubra, clade e—Yonagunia, clade f—Dermocorynus, and clade g—Mariaramirezia). The monophyly of Grateloupia sensu stricto (69% MLB) and Phyllymenia (30% MLB) was not strongly supported. In addition to the named genera, three other groups of species were fully supported (clades c, d, and h). In the genus-level relationships within the overall Halymeniales, the sister relationship (95% MLB) of two monophyletic genera, Polyopes (99% MLB) and Glaphyrosiphon (100% MLB), was the only supported clade in the rbcL tree.
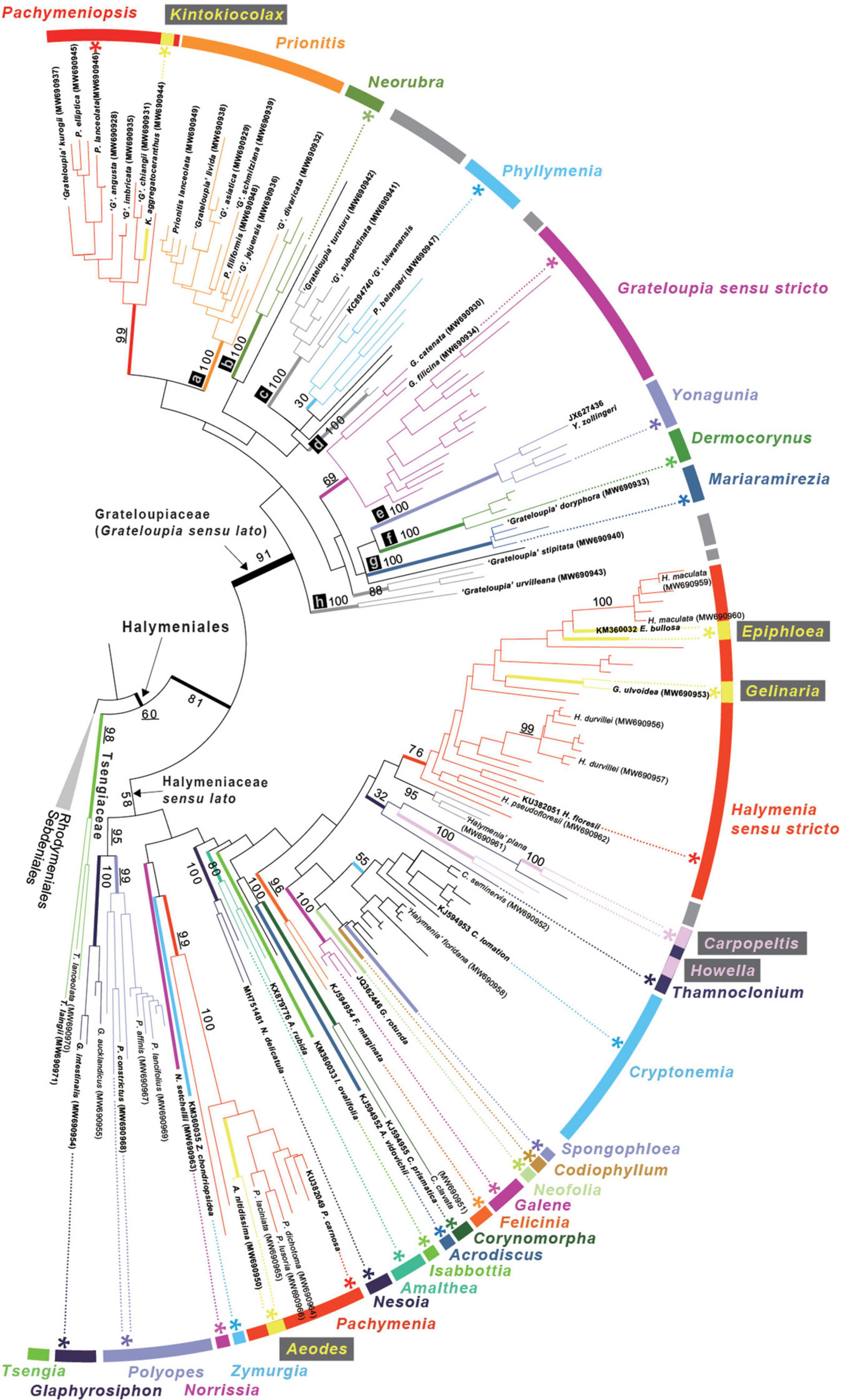
Figure 2. ML phylogeny of the Halymeniales using rbcL. Maximum Likelihood (ML) phylogeny of Halymeniales inferred from 207 rbcL sequences. The tips with GenBank accessions in parentheses indicate newly analyzed rbcL sequences in this study. Bolded taxa indicate the species selected for 5-gene analyses. Numbers at bold nodes correspond to bootstrap support for some important nodes. Asterisk (*) indicates type species of each genus. Labeled tree with all taxon names and bootstrap support is shown in Supplementary Figure 1.
Five gene sequences, rbcL, psaA, psbA, cox1, and LSU from 47 taxa were used to infer the individual gene phylogenies of the Halymeniales (Supplementary Table 2). Individual gene analyses yielded a generally poorly resolved tree (Supplementary Figures 2–5). The monophyly of Grateloupia sensu lato was strongly supported in the rbcL, psaA and LSU trees (91, 99, and 100% MLB; Supplementary Figures 1,2,5). However, support for the monophyly of Yonagunia with other species of Grateloupia sensu lato in psbA and cox1 analyses was absent or weak (Supplementary Figures 3, 4).
A five-gene concatenated data set was used to improve the resolution of the phylogeny (i.e., relationships and support for monophyly) within the Halymeniales (Figure 3). The concatenated tree was mostly concordant with the rbcL tree and provided better resolution at the genus, family, and ordinal levels compared to individual trees (Figure 3 and Supplementary Figures 2–5). The monophyly of Grateloupia sensu lato and of the Halymeniales were fully supported in the concatenated analyses (100% MLB for both nodes). The five-gene combined tree showed that the Tsengiaceae diverged first (100% MLB), followed by the monophyletic clade consisting of Pachymenia and Aeodes (83% MLB). After the divergence of a Pachymenia-Aeodes clade, Zymurgia, Norrissia, and Isabbottia diverged sequentially. After diverging from Isabbottia, 12 genera grouped with low support (44% MLB). Within this clade, Halymenia, the type genus of the Halymeniaceae, Epiphloea and Gelinaria were grouped as a monophyletic clade with strong support (96% MLB). Polyopes and Glaphyrosiphon show a sister relationship with strong support (100% MLB). The Grateloupia sensu lato clade was monophyletic with strong support (100% MLB) and comprised of Pachymeniopsis, Kintokiocolax, Prionitis, Phyllymenia, Grateloupia, and Yonagunia. Within the Grateloupia sensu lato, Yonagunia was resolved at a basal position (100% MLB).
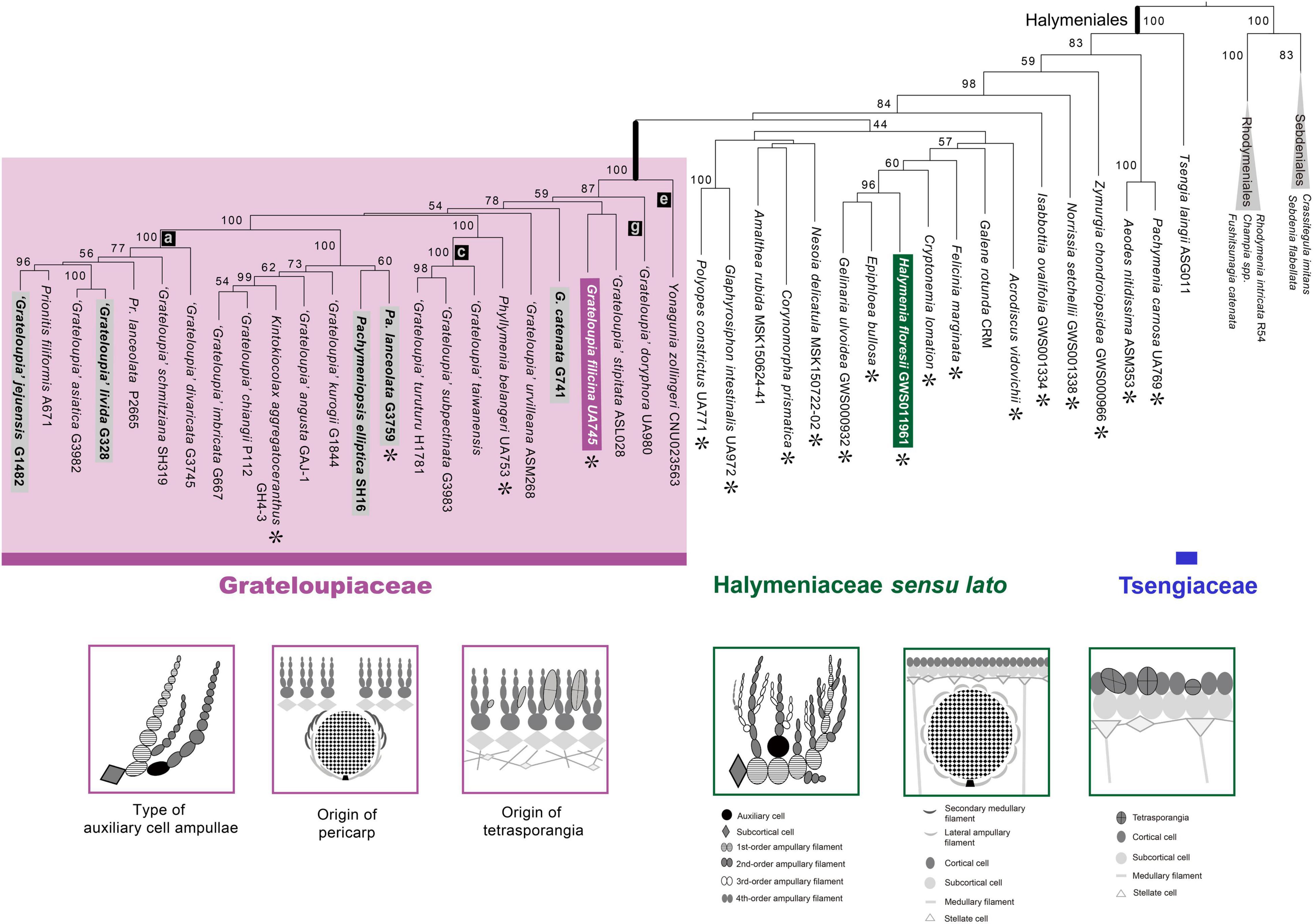
Figure 3. ML phylogeny of the Halymeniales using a five-gene combined data set with three distinguishing morphological characteristics of the Grateloupiaceae compared to Halymenia floresii. The ML phylogeny was constructed using a five-gene concatenated data set of rbcL, psaA, psbA, cox1, and LSU. Purple colored boxes indicate the characteristics of the Grateloupiaceae, and green colors show the characteristics of Halymenia floresii, the type species of the Halymeniaceae. Asterisks (∗) indicate the type species of each genus. Gray boxes indicate the taxa used for morphological observations.
Morphological Observation
We observed the morphology of the reproductive structures of five representative species from three different genera in Grateloupia sensu lato, focusing on the following characteristics: (i) type of auxiliary cell ampullae, (ii) origin of the pericarp, and (iii) tetrasporangial development.
The type of auxiliary cell ampullae in Grateloupia sensu lato was the simple lateral branched type and initiated from the subcortical cell having two or sometimes three orders of ampullary filaments laterally branched from the first-order ampullary filament (Figures 4A–C). The auxiliary cell ampullae of Grateloupia catenata are three-ordered, which has a second-order ampullary filament arising laterally from the second cell of the first-order ampullary filament (Figure 4A). The auxiliary cell is the basal cell of a second-order filament produced on the second or third cell of the first-order filament with a cell considered as a basal cell of a third order filament (Figure 4A). The number of cells of each ampullary filament is more than nine cells long in the first order filaments and more than eight cells long in the second order filaments (Figure 4A). In “Grateloupia” jejuensis, the auxiliary cell ampullae, which are two-ordered, originate from a subcortical cell (Figure 4B). Three second-order ampullary filaments arise laterally from the first, the second, and the third cell of the first-order ampullary filament (Figure 4B). The auxiliary cell is positioned on the third cell of a first order filament which is composed of more than five cells long (Figure 4B) and is the basal cell of second-order filament which is composed of more than seven cells in length (Figure 4B). The fully developed auxiliary cell ampullae is observed in Pachymeniopsis elliptica having distinctly two-ordered ampullary filaments, with the first-order filament composed of more than 11 cells in length and the second comprised of up to twenty-one cells long laterally (Figure 4C). The first cell of the second-order ampullary filament enlarges and functions as the auxiliary cell (Figure 4C).
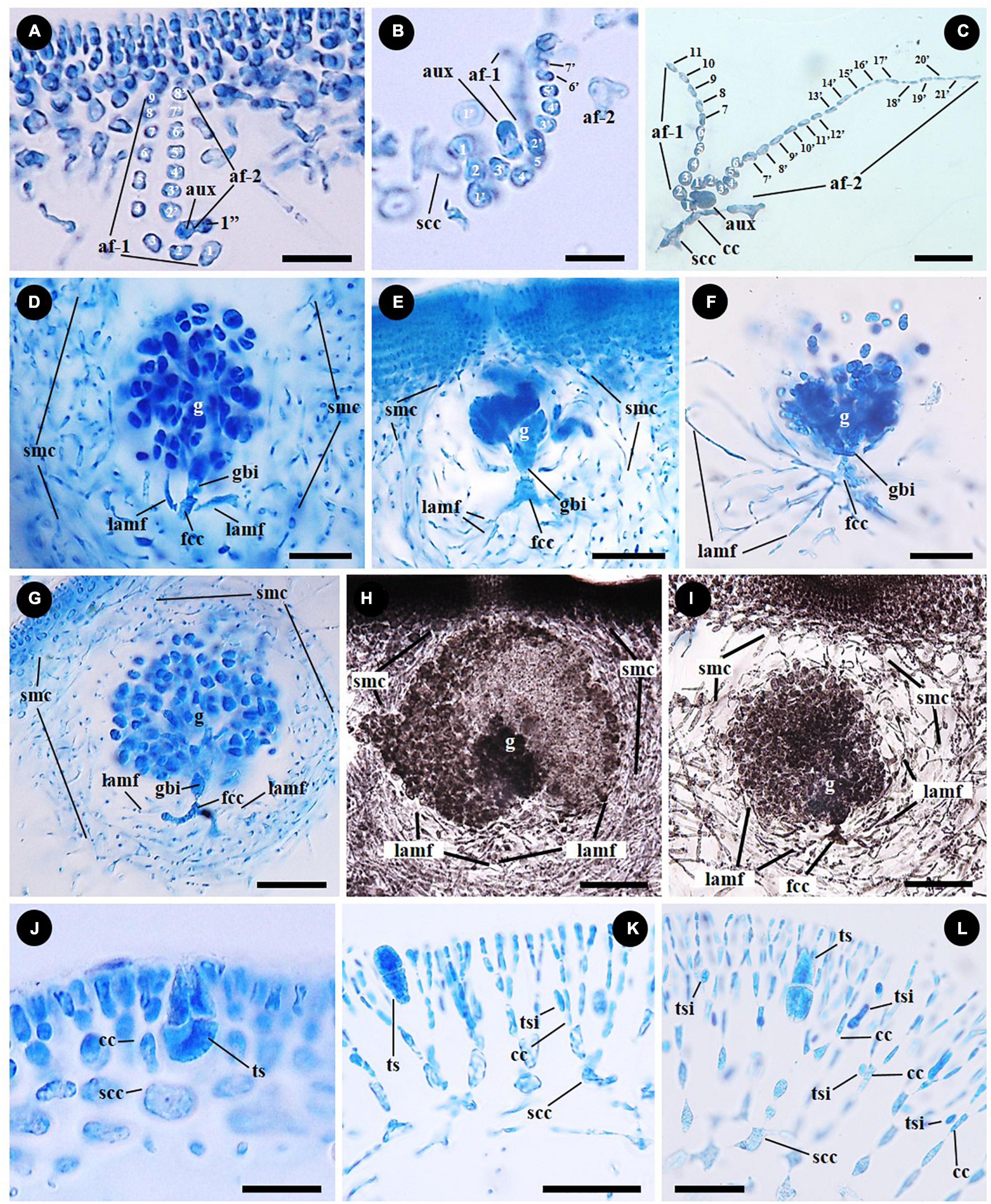
Figure 4. Female reproductive structures and tetrasporangial development of representative species of the Grateloupiaceae: Grateloupia catenata (A,D,G,J), “Grateloupia” jejuensis (B,E), “G.” livida (H,K), Pachymeniopsis elliptica (C,F), and Pa. lanceolata (I,L). (A–C) 2-ordered auxiliary cell ampullae originated from subcortical cell (scc) or inner cortical cell (cc) of the representative species G. catenata (A, MSK180814-01), “G.” jejuensis (B, MSK170722-32) and Pa. elliptica (C, MSK170417-57). Each auxiliary cell (aux) is the basal cell of a second order ampullary filament (af-2) borne from a first order cell (af-1). (D–F) Young gonimoblasts originated from fusion cell complex (fcc), the gonimoblast initial (gbi) and the elongation and branching of lateral ampullary filaments in the gonimolobe (g) in G. catenata (D, MSK180814-01), “G.” jejuensis (E, MSK170722-37) and Pa. elliptica (F, MSK170417-58). Lateral ampullary filaments (lamf) and secondary medullary cells (smc). (G–I) Pericarp composed of lateral ampullary filaments of a fusion cell complex (fcc) and secondary medullary filaments (smc) originated from the subcortex in G. catenata (G, MSK180814-01), “G.” livida (H, MSK170417-67) and Pa. lanceolata (I, MSK150502-05). Laterally entangled ampullary filaments and numerous secondary medullary filaments surrounding the gonimolobe (g). (J–L) Tetrasporangia (ts) cruciately divided, and tetrasporangial initial (tsi) originated unilaterally from inner cortical cell (cc) in G. catenata (J, MSK180814-02), “G.” livida (K, MSK140224-22) and Pa. lanceolata (L, MSK161126-01). Scale bars: (A,B) = 15 μm; (C,D,L) = 25 μm; (E–G,I,K) = 50 μm; (H) = 100 μm; (J) = 20 μm.
The female reproductive developmental stages after fertilization are similar in G. catenata, a member of Grateloupia sensu stricto, “G.” jejuensis and “G.” livida of Prionitis, and Pa. lanceolata, the generitype of Pachymeniopsis with Pa. elliptica. The auxiliary cell and basal auxiliary cell ampullary filaments become incorporated into a fusion cell complex (marked as “fcc” in Figures 4D–G). The ampullary filaments of a fusion cell complex each grow as lateral filaments (marked as “lamf” in Figures 4D–I), and the gonimoblast initial is cut off from the fusion cell complex by a transverse division (Figures 4D–F). The gonimoblast initial, which is concave to spherical in shape, is divided transversely and vertically. During carposporophyte development, the lateral ampullary filaments produced from the fusion cell complex are growing up longitudinally and are repeatedly dichotomously branched (Figures 4D–F). The secondary medullary filaments issued from the subcortex and the lateral ampullary branches surround the base of the gonimolobe (Figures 4D–F) and the pericarp is formed by the linking of secondary medullary filaments (marked as “smc” in Figures 4G–I) produced from subcortical cells and of lateral ampullary filaments borne from a fusion cell complex during the gonimoblast initiation stage (Figures 4G–I).
Tetrasporophytes are isomorphic with the gametophytes in all species. Tetrasporangia are usually scattered over the entire thallus. Tetrasporangia are initiated laterally from the cell files of the outer cortex: the basal cell of cortical cell file in G. catenata (Figure 4J); the fourth cell layer from the surface in “G.” livida (Figure 4K); the third to fifth cell layer from the surface in Pa. lanceolata (Figure 4L). Tetrasporangial initials are globular to kidney shaped, and narrowly elongated (Figures 4K,L). The tetrasporangia are divided transversely at first (Figure 4K), and then longitudinally (Figure 4L). The mature tetrasporangia are ellipsoidal and positioned between dichotomously branched cortical filaments (Figures 4J–L).
Taxonomic Treatment
Grateloupiaceae Schmitz (1889) emend. S. Y. Kim, H. W. Lee, and M. S. Kim
Description: Thallus terete or decumbent, multiaxial, angular, flat to leaf-like, compressed, variably forked, or usually branched laterally, in the majority of cases with a very clear filiform medullary structure; carpogonia and auxiliary cells scattered individually; carpogonial branches 2-celled on carpogonial branch ampullae; auxiliary cell ampullary filaments unilaterally unbranched, pericarp formed by the fusion of secondary medullary filaments from subcortical cells and lateral ampullary filaments from a fusion cell complex; tetrasporangia ellipsoidal, arising laterally from inner cells of outer cortex.
Type genus: Grateloupia C. Agardh, 1822
Included genera: Grateloupia C. Agardh, Dermocorynus P. Crouan, and H. Crouan, Prionitis J. Agardh, Pachymeniopsis Yamada ex Kawabata, Kintokiocolax Tak.Tanaka and Y. Nozawa, Phyllymenia J. Agardh, Neorubra M. S. Calderon, G. H. Boo, and S. M. Boo, Mariaramirezia M. S. Calderon, G. H. Boo, A. Mansilla and S. M. Boo and Yonagunia S. Kawaguchi and M. Masuda.
Discussion
We consider that the resurrection of the monophyletic Grateloupiaceae is a first step toward resolving the Halymeniales with the aim of adapting the classification system based on genetic monophyly. In addition to all available rbcL sequences from previous studies (e.g., Wang et al., 2001; Gargiulo et al., 2013; Calderon et al., 2014a, b), we determined 45 new rbcL sequences. Our rbcL data set covers more than 60% (82 species out of 138) of currently described species from Grateloupiaceae. On the other hand, it is noteworthy that the genera of the Halymeniales, except for the Grateloupiaceae and Tsengiaceae, were not resolved as a monophyletic lineage in the present and previous analyses (Manghisi et al., 2014, 2017). We believe that this is due to insufficient taxon sampling from Halymenia-Cryptonemia related genera caused by two reasons: known species that are not analyzed and taxa that are not yet known. Although Cryptonemia (51 spp.) and Halymenia (78 spp.) account for more than 30% within the Halymeniales (358 spp.) and are the second and third largest genera in the order, few studies have been conducted on these genera compared to Grateloupia. In addition, recent studies have continuously discovered new taxa within this group (e.g., Rodríguez-Prieto et al., 2018; Lee and Kim, 2019; Schneider et al., 2019). We expected that extensive studies using taxon-rich sampling focused on Halymenia-Cryptonemia and related genera could resolve the non-monophyly of the Halymeniaceae, but this is beyond the scope of this study. At this point, we tentatively used the “Halymeniaceae sensu lato” for the genera of Halymeniales except for the Grateloupiaceae and Tsengiaceae (Figures 2, 3). A more comprehensive study on the Halymeniaceae sensu lato is pending in a future study.
There have been no extensive molecular phylogenetic studies conducted of the Halymeniales as a whole so far. However, Wang et al. (2001) discussed the phylogeny of Halymeniales based on rbcL sequence analyses. They suggested the Pachymenia-Aeodes clade was basal within the Halymeniaceae followed by the Cryptonemia-Halymenia complex clade, a Polyopes clade, then a Grateloupia-Prionitis clade. This was the first extensive study for the Halymeniales; however, they rather focused on few specific genera (e.g., Prionitis). Although these supergeneric clades have been recovered in subsequent systematic studies on the Halymeniaceae (e.g., Nelson et al., 2014; Manghisi et al., 2017), those studies were based on a limited number of genera, and the Tsengiaceae was used as an outgroup or was not included. Meanwhile, Saunders and colleagues suggested several familial and ordinal level changes based on limited genera (Figures 1A–C). To overcome limitations in previous studies, we extensively included 33 genera out of 38 with 30 type species of the Halymeniales except for five genera of Halymeniaceae. The taxonomic status of these five genera are unstable (Figure 1). For instance, the monotypic genera Blastophye, Hymenophlea, and Zanardinula have been regarded as a synonym of Prionitis, Neoabbotiella, which has been referred to as being part of Cryptonemia (Guiry and Guiry, 2021), and Grateloupiocolax, which has never been studied after the first description from the Caribbean coast (Schnetter et al., 1983).
Based on this taxon-rich rbcL phylogeny, we designated the representative taxa and performed a multi-gene phylogeny of the Halymeniales. Phylogenetic analyses of a five-gene combined data set have shown that the Tsengiaceae diverged first (100% MLB; Figure 3) within the Halymeniales followed by Pachymenia and Aeodes. The basal position of Pachymenia and Aeodes within the Halymeniaceae is largely congruent with the phylogenetic concepts of Chiang (1970) and Fredericq et al. (1996) based on reproductive morphology, as well as previous molecular phylogenies (e.g., Wang et al., 2001). After the divergence of Pachymenia and Aeodes, three monotypic genera have distinct phylogenetic positions (Isabbottia, Norrissia, and Zymurgia; Figure 3). Among the other genera of Halymeniaceae sensu lato, the sister relationship of Polyopes with Glaphyrosiphon, which have been suggested as a sister taxon in previous studies (Hommersand et al., 2010; Manghisi et al., 2014), was fully supported (100% BTS). Lastly, the Grateloupiaceae was recovered as a monophyletic clade with full support (100% MLB; Figure 3). The close relationships among the species of the Grateloupiaceae and their differences to the other species of Halymeniaceae were also supported by several morphological characteristics in common regarding female and tetrasporangial developments: type of auxiliary cell ampullae (Chiang, 1970), pericarp origin (Balakrishnan, 1961a, b), and origin of tetrasporangia (Figures 3, 4).
The members of the Halymeniaceae and Grateloupiaceae share the following reproductive features: 2-celled carpogonial branches, similar ampullae structures producing carpogonial branches and auxiliary cells, formation of a pericarp, and cruciately divided tetrasporangia (Chiang, 1970; Womersley and Lewis, 1994; Rodríguez-Prieto et al., 2018). The monogeneric family Tsengiaceae has no auxiliary cell ampullae (Fan and Fan, 1962), which makes it distinct from the Grateloupiaceae and Halymeniaceae. The auxiliary cell ampullae of the Grateloupiaceae consist of two-or three-ordered ampullary filaments mostly having one or two, sometimes 3 s-order ampullary filaments branched laterally from the first-order ampullary filament (Figures 4A–C; Kawabata, 1962, 1963; Kawaguchi, 1989; Kawaguchi et al., 2004; Lin et al., 2008). Although there are different interpretations for the development of auxiliary cell ampullae in some species groups of Grateloupiaceae (Gargiulo et al., 2013; Calderon et al., 2014a, b), we consider that the type of auxiliary cell ampullae present in the Grateloupiaceae is of the simple lateral branched type regardless of the number of ampullary cell orders and the position of the auxiliary cell, a situation that can be distinguished from the other halymeniacean taxa. In the case of Halymenia floresii, the species has the Halymenia-type auxiliary cell ampullae, 3–4-ordered auxiliary cell ampullae producing two and more second-order ampullary filaments laterally from the first-order ampullary filament, with several short third-order ampullary filaments (Chiang, 1970; Rodríguez-Prieto et al., 2018). Compared to other members of the Halymeniaceae such as Aeodes, Cryptonemia, Glaphyrosiphon, Pachymenia, Polyopes, and Thamnoclonium, the auxiliary cell ampullae of the Grateloupiaceae are distinctly simple enough to be distinguished phylogenetically (Table 1).
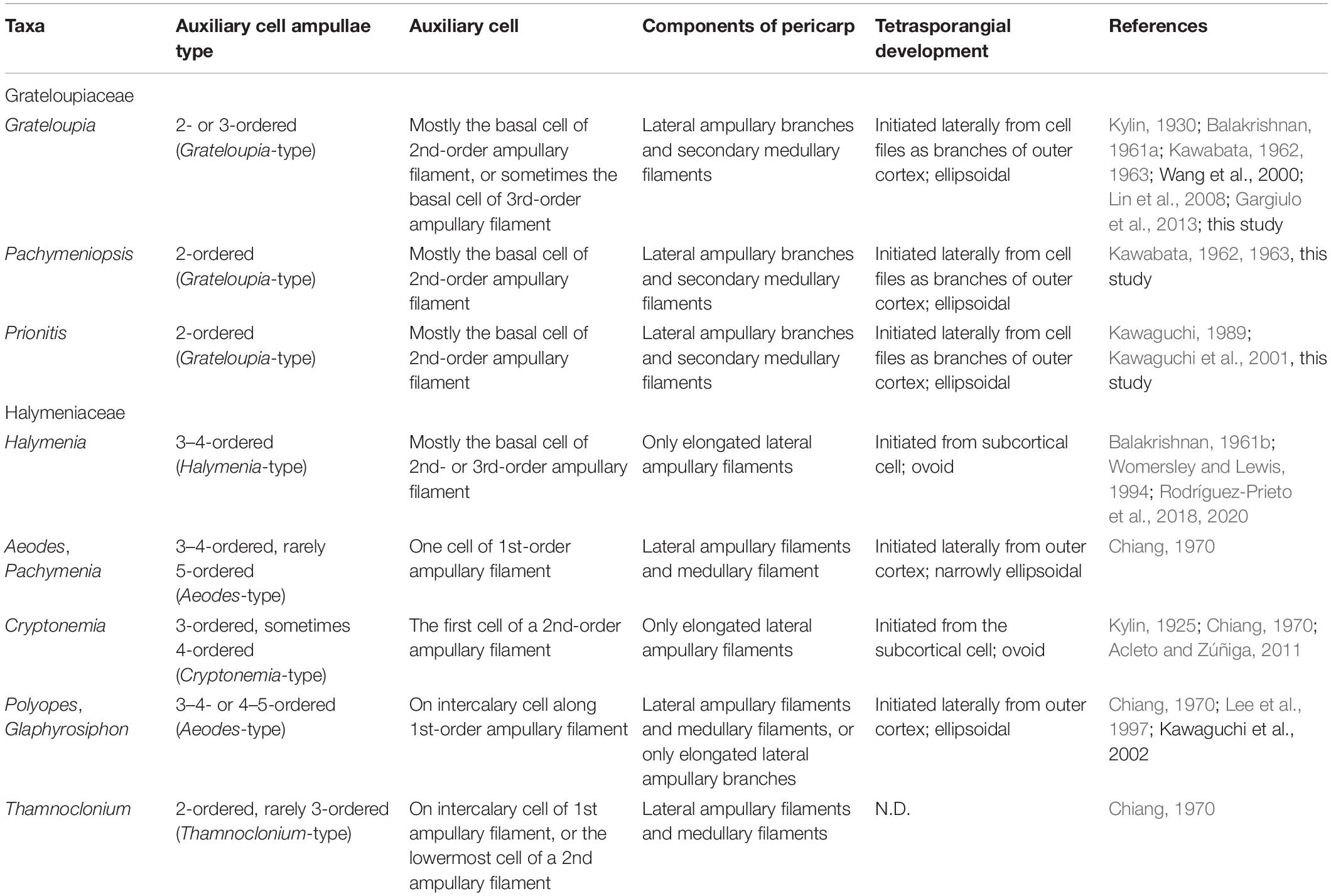
Table 1. The comparison of the reproductive structures of the representative members of Grateloupiaceae and Halymeniaceae, respectively.
The pericarps of the Grateloupiaceae are formed by the fusion of secondary medullary filaments which are initiated from subcortical cells, and of lateral ampullary filaments borne from a fusion cell complex (Figures 4G–I; Balakrishnan, 1961a; Kawabata, 1962, 1963; Kawaguchi, 1989; Lin et al., 2008; Gargiulo et al., 2013). However, although the pericarps of Codiophyllum, Corynomorpha, Pachymenia, Polyopes, and Thamnoclonium, taxa belonging to the Halymeniaceae clade (Figure 2), have been described as formed by elongated ampullary filaments and neighboring medullary filaments (Balakrishnan, 1962; Chiang, 1970; Womersley and Lewis, 1994; Kawaguchi et al., 2002), their involucral networks are looser than those of the Grateloupiaceae. In the members of Cryptonemia, the pericarps are mostly consisting of elongated lateral ampullary filaments, or sometimes of lateral ampullary filaments and neighboring medullary filaments (Kylin, 1925; Chiang, 1970; Lewis, 1994; Acleto and Zúñiga, 2011). The pericarps of the members of Halymenia are produced only by ampullary filaments originating from a fusion cell complex (Rodríguez-Prieto et al., 2018). In other Halymeniaceae sensu lato taxa such as Amalthea, Galene, Glaphyrosiphon, Neofolia, and Nesoia, the pericarps are produced by elongated lateral ampullary filaments only (Hommersand et al., 2010; D’Archino et al., 2014; Rodríguez-Prieto et al., 2018, 2019, 2020; Lee and Kim, 2019).
Tetrasporangia in the Grateloupiaceae are initiated laterally from inner cells of the outer cortex and grow outwardly between dichotomously branched outer cortical filaments such as the other cell files of the outer cortex (Figures 4J–L). Regarding tetrasporangia, type of division (i.e., cruciate or tetrahedral), shape, and size have been used to describe species and classify halymeniacean taxa (e.g., Kawaguchi et al., 2001; Saunders and Kraft, 2002; Rodríguez-Prieto et al., 2018). However, in our study we focused on the initiation of tetrasporangia. The tetrasporangia of Halymenia including Halymenia sensu stricto and Cryptonemia members are ovoid and similar in size with the outermost cortical cells produced from subcortical cells (Chiang, 1970; D’Archino et al., 2014; Rodríguez-Prieto et al., 2018, 2019, 2020; Lee and Kim, 2019). The division type of tetrasporangia is one of the diagnostic characters classifying the family groups in red algae (Kylin, 1932). Although tetrasporangial division is cruciate in all members of Grateloupiaceae and Halymeniaceae (Chiang, 1970; Womersley and Lewis, 1994), the tetrasporangia of the Grateloupiaceae are ellipsoidal, and initiated laterally from the third to the fifth cell, up to the sixth, of outer cortical cell files beneath the thallus surface, whereas their initiation in each group in the Halymeniaceae is different: in Halymenia, including Halymenia sensu stricto, and Cryptonemia, they are ovoid, initiated from a subcortical cell; in Aeodes, Pachymenia, Polyopes, and Glaphyrosiphon, they are ellipsoidal, initiated laterally from a cortical cell (Table 1).
Considering our phylogenetic results and morphological observations on reproductive structures, including the taxonomic history (Wang et al., 2001; Gargiulo et al., 2013), the monophyletic Grateloupia sensu lato clade is clearly distinct from the representative halymeniacean taxa including Halymenia floresii. As a result, we reinstate the family Grateloupiaceae based on the generitype Grateloupia, which was established by Schmitz (1889), but later synonymized with the Halymeniaceae (as Cryptonemiaceae) by Papenfuss (1955). Consequently, we propose the resurrection of the family name Grateloupiaceae Schmitz for the Grateloupia sensu lato clade as a distinct family within the Halymeniales and also provide an emended genus composition. Based on our current study, the emended Grateloupiaceae now includes Grateloupia, Dermocorynus, and Prionitis among the 11 genera of Grateloupiaceae sensu Schmitz, 1889 and an additional six genera, Pachymeniopsis, Phyllymenia, Neorubra, Mariaramirezia, Kintokiocolax, and Yonagunia. Although Dermocorynus, Mariaramirezia, and Neorubra were not included in our multigene analyses, we consider them as members of the Grateloupiaceae based on the rbcL phylogeny and previous studies (Wilkes et al., 2005; Calderon et al., 2014a, b).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: MW690928–MW691096).
Author Contributions
HSY and MSK designed the study. SYK and ECY performed molecular experiment, data analyses, and drafted the molecular section. HWL conducted morphological observation and drafted the morphological section. SMB, JL-B, SF, and RD’A contributed the samples collection and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was partly supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2017R1A2B3001923, 2019R1I1A1A01058544, 2019R1A6A1A10072987, 2020R1I1A2069706, and 2021R1I1A1A010 44280) and funded by the Ministry of Science and ICT (NRF-2019R1A2C2088565), and by the U.S. National Science Foundation (0936884, 1317114, and 0937978). Collections from New Zealand were supported by the NIWA SSIF funding to the Marine Biological Resources Program, Coasts and Oceans National Center.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We gratefully acknowledge Gary W. Saunders and Wendy A. Nelson for collecting precious algal samples and allowing their use for this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.775627/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov
- ^ https://v3.boldsystems.org
- ^ http://www.technelysium.com.au/chromas.html
References
Acleto, C., and Zúñiga, R. (2011). Revisión de las especies peruanas de Sebdenia (Sebdeniales, Rhodophyta) y descripción de Cryptonemia anconensis sp. nov. (Halymeniales, Rhodophyta). Rev. Peru. Biol. 18, 97–112.
Balakrishnan, M. S. (1961a). Studies on Indian Cryptonemiales–I. Grateloupia C. A. Ag. J. Madras Univ. 31B, 11–35.
Balakrishnan, M. S. (1961b). Studies on Indian Cryptonemiales–III. Halymenia C. A. Ag. J. Madras Univ. 31B, 183–217.
Balakrishnan, M. S. (1962). Studies on Indian Cryptonemiales II. Corynomorpha. J. Ag. Phytomorphol. 12, 77–86.
Boo, G. H., Le Gall, L., Miller, K. A., Freshwater, D. W., Wernberg, T., Terada, R., et al. (2016). A novel phylogeny of the Gelidiales (Rhodophyta) based on five genes including the nuclear cesA, with descriptions of Orthogonacladia gen. nov. and Orthogonacladiaceae fam. nov. Mol. Phylogenet. Evol. 101, 359–372. doi: 10.1016/j.ympev.2016.05.018
Bory de Saint-Vincent, J. B. (1828). “Botanique, cryptogamie,” in Voyage Autour Du Monde, Executé Par Ordre Du Roi, Sur La Corvette De Sa Majesté, la Coquille, Pendant Les Années 1822, 1823, 1824 et 1825, ed. L. I. Duperrey (Paris: Bertrand), 178–181.
Calderon, M. S., Boo, G. H., and Boo, S. M. (2014a). Morphology and phylogeny of Ramirezia osornoensis gen. & sp. nov. and Phyllymenia acletoi sp. nov. (Halymeniales, Rhodophyta) from South America. Phycologia 53, 23–36. doi: 10.2216/13-158.1
Calderon, M. S., Boo, G. H., and Boo, S. M. (2014b). Neorubra decipiens gen. & comb. nov. and Phyllymenia lancifolia comb. nov. (Halymeniales, Rhodophyta) from South America. Phycologia 53, 409–422. doi: 10.2216/14-027.1
Calderon, M. S., Boo, G. H., and Boo, S. M. (2016). Correction to the paper “Morphology and phylogeny of Ramirezia osornoensis gen. & sp. nov. and Phyllymenia acletoi sp. nov. (Halymeniales, Rhodophyta) from South America”. Phycologia 55:610.
Chiang, Y.-M. (1970). Morphological studies of red algae of the family Cryptonemiaceae. Univ. Calif. Publ. Bot. 58, 1–95.
D’Archino, R., Nelson, W. A., and Zuccarello, G. C. (2014). Amalthea and Galene, two genera of Halymeniaceae (Rhodophyta) from New Zealand. Bot. Mar. 57, 185–201. doi: 10.1515/bot-2014-0008
De Clerck, O., Gavio, B., Fredericq, S., Bárbara, I., and Coppejans, E. (2005a). Systematics of Grateloupia filicina (Halymeniaceae, Rhodophyta), based on rbcL sequence analyses and morphological evidence, including the reinstatement of G. minima and the description of G. capensis sp. nov. J. Phycol. 41, 391–410. doi: 10.1111/j.1529-8817.2005.04189.x
De Clerck, O., Gavio, B., Fredericq, S., Cocquyt, E., and Coppejans, E. (2005b). Systematic reassessment of the red algal genus Phyllymenia (Halymeniaceae, Rhodophyta). Eur. J. Phycol. 40, 169–178. doi: 10.1080/09670260500128343
Dixon, K. R., Saunders, G. W., Schneider, C. W., and Lane, C. E. (2015). Etheliaceae fam. nov. (Gigartinales, Rhodophyta), with a clarification of the generitype of Ethelia and the addition of six novel species from warm waters. J. Phycol. 51, 1158–1171. doi: 10.1111/jpy.12353
Entwisle, T. J., Evans, J. R., Vis, M. L., and Saunders, G. W. (2018). Ottia meiospora (Ottiaceae, Rhodophyta), new genus and family endophytic within the thallus of Nothocladus (Batrachospermales, Rhodophyta). J. Phycol. 54, 79–84. doi: 10.1111/jpy.12603
Fan, K. C., and Fan, Y. P. (1962). Studies on the reproductive organs of red algae I. Tsengia and the development of its reproductive systems. Acta Bot. Sin. 10, 187–196.
Fredericq, S., Hommersand, M. H., and Freshwater, D. W. (1996). The molecular systematics of some agar- and carrageenan-containing marine red algae based on rbcL sequence analysis. Hydrobiologia 32, 125–136. doi: 10.1007/BF00047797
Freshwater, D. W., Fredericq, S., and Bailey, J. C. (1999). Characteristics and utility of nuclear-encoded large-subunit ribosomal gene sequences in phylogenetic studies of red algae. Phycol. Res. 47, 33–38. doi: 10.1111/j.1440-1835.1999.tb00281.x
Gargiulo, G. M., Morabito, M., and Manghisi, A. (2013). A re-assessment of reproductive anatomy and postfertilization development in the systematics of Grateloupia (Halymeniales, Rhodophyta). Crypt. Algol. 34, 3–35. doi: 10.7872/crya.v34.iss1.2013.3
Gavio, B., Hickerson, E., and Fredericq, S. (2005). Platoma chrysymenioides sp. nov. (Schizymeniaceae), and Sebdenia integra sp. nov. (Sebdeniaceae), two new red algal species from the northwestern Gulf of Mexico, with a phylogenetic assessment of the Cryptonemiales complex (Rhodophyta). Gulf Mexico Sci. 2005, 38–57. doi: 10.18785/goms.2301.05
Guiry, M. D. (1978). Notes on some family names of Florideophyceae (Rhodophyta). Taxon 27, 191–195. doi: 10.2307/1220239
Guiry, M. D., and Guiry, G. M. (2021). AlgaeBase. Galway: World-wide electronic publication, National University of Ireland.
Guiry, M. D., and Maggs, C. A. (1982). The morphology and life history of Dermocorynus montagnei Crouan frat. (Halymeniaceae; Rhodophyta) from Ireland. Br. Phycol. J. 17, 215–228. doi: 10.1080/00071618200650231
Harvey, W. H. (1849). A Manual of the British Marine Algae: Containing Generic and Specific Descriptions of All the Known British Species of Sea-Weeds. With Plates to Illustrate all the Genera. London: John Van Voorst, Paternoster Row, 121–252. doi: 10.5962/bhl.title.152174
Hommersand, M. H., and Fredericq, S. (1990). “Sexual reproduction and cystocarp development,” in Biology of the Red Algae, eds K. M. Cole and R. G. Sheath (Cambridge, MA: Cambridge University Press), 305–345.
Kawabata, S. (1962). A contribution to the systematic study of Grateloupiaceae from Japan (1). Mem. Hokkaido Gakugei Univ. 13, 22–51.
Kawabata, S. (1963). A contribution to the systematic study of Grateloupiaceae from Japan (2). Mem. Hokkaido Gakugei Univ. 13, 190–210.
Kawaguchi, S. (1989). The genus Prionitis (Halymeniaceae, Rhodophyta) in Japan. J. fac. Sci. Hokkaido Univ. Ser. V 14, 193–257.
Kawaguchi, S., Shimada, S., Wang, H. W., and Masuda, M. (2004). The new genus Yonagunia Kawaguchi et Masuda (Halymeniaceae, Rhodophyta), based on Y. tenuifolia Kawaguchi et Masuda sp. nov. from southern Japan and including Y. formosana (Okamura) Kawaguchi et Masuda comb. nov. from southeast Asia. J. Phycol. 40, 180–192. doi: 10.1046/j.1529-8817.2004.03077.x
Kawaguchi, S., Wang, H. W., Horiguchi, T., Sartoni, G., and Masuda, M. (2001). A comparative study of the red alga Grateloupia filicina (Halymeniaceae) from the northwestern Pacific and Mediterranean with the description of Grateloupia asiatica sp. nov. J. Phycol. 37, 433–442. doi: 10.1046/j.1529-8817.2001.037003433.x
Kim, M. S., Kim, S. Y., and Nelson, W. (2010). Symphyocladia lithophila sp. nov. (Rhodomelaceae, Ceramiales), a new Korean red algal species based on morphology and rbcL sequences. Bot. Mar. 53, 233–241. doi: 10.1515/BOT.2010.031
Krayesky, D. M., Norris, J. N., Gabrielson, P. W., Gabriela, D., and Fredericq, S. (2009). A new order of red algae based on the Peyssonneliaceae, with an evaluation of the ordinal classification of the Florideophycae (Rhodophyta). Proc. Biol. Soc. Washington 122, 364–391. doi: 10.2988/08-43.1
Kylin, H. (1925). The marine red algae in the vicinity of the Biological Station at Friday Harbor, Wash. Linds Univ. Årsskrift Ny Följd Andra Afdelningen 21, 1–87.
Le Gall, L., and Saunders, G. W. (2007). A nuclear phylogeny of the Florideophyceae (Rhodophyta) inferred from combined EF2, small subunit and large subunit ribosomal DNA : establishing the new red algal subclass Corallinophycidae. Mol. Phylogenet. Evol. 43, 1118–1130. doi: 10.1016/j.ympev.2006.11.012
Lee, H. W., and Kim, M. S. (2019). Female reproductive structures define the novel genus, Nesoia (Halymeniaceae, Rhodophyta). Eur. J. Phycol. 54, 66–77. doi: 10.1080/09670262.2018.1513166
Lee, H. B., Lewis, J. A., Kraft, G. T., and Lee, I. K. (1997). Sinkoraena gen. nov. (Halymeniaceae, Rhodophyta) from Korea, Japan and southern Australia. Phycologia 36, 103–113.
Lewis, J. A. (1994). Transfer of the Australian red algae Kallymenia nitophylloides to Cryptonemia (Halymeniaceae) and Halymenia chondricola to Hymenocladia (Rhodymeniaceae). Taxon 43, 3–10.
Lin, S.-M., D’Archino, R., and Hommersand, M. H. (2012). A new method of cystocarp development in the red algal genus Callophyllis (Kallymeniaceae) from Chile. J. Phycol. 48, 784–792. doi: 10.1111/j.1529-8817.2012.01151.x
Lin, S.-M., Liang, H.-Y., and Hommersand, M. H. (2008). Two types of auxiliary cell ampullae in Grateloupia (Halymeniaceae, Rhodophyta), including G. taiwanensis sp. nov. and G. orientalis sp. nov. from Taiwan based on rbcL gene sequence analysis and cystocarp development. J. Phycol. 44, 196–214. doi: 10.1111/j.1529-8817.2007.00443.x
Manghisi, A., Le Gall, L., Bonillo, C., Gargiulo, G. M., Ribera, M. A., and Morabito, M. (2017). An assessment of the taxonomic status of the Mediterranean endemic genus Acrodiscus zanardini (Halymeniales, Rhodophyta). Eur. J. Taxon. 267, 1–24. doi: 10.5852/ejt.2017.267
Manghisi, A., Le Gall, L., Ribera, M. A., Bonillo, C., Gargiulo, G. M., and Morabito, M. (2014). The Mediterranean endemic new genus Felicinia (Halymeniales, Rhodophyta) recognized by a morphological and phylogenetic integrative approach. Crypt. Algol. 35, 221–243. doi: 10.7872/crya.v35.iss3.2014.221
Nelson, W. A., Kim, S. Y., and Boo, S. M. (2014). Transfer of the subantarctic red alga Grateloupia aucklandica to the genus Glaphyrosiphon (Halymeniales, Rhodophyta). Phycologia 53, 457–462. doi: 10.2216/14-017.1
Papenfuss, G. F. (1955). “Classification of the algae,” in A Century of Progress in the Natural Science, 1853-1953, ed. E. L. Kessel (San Francisco, CA: California Academy of Sciences), 115–224.
Rambaut, A. (2009). FigTree v1.3.1: Tree Figure Drawing Tool. Available online at: http://tree.bio.ed.ac.uk/software/figtree/
Rodríguez-Prieto, C., De Clerck, O., Huisman, J. M., and Lin, S.-M. (2018). Systematics of the red algal genus Halymenia (Halymeniaceae, Rhodophyta): characterization of the generitype H. floresii and description of Neofolia rosea gen. et sp. nov. Eur. J. Phycol. 53, 520–536. doi: 10.1080/09670262.2018.1478132
Rodríguez-Prieto, C., De Clerck, O., Huisman, J. M., and Lin, S.-M. (2019). Characterisation of Nesoia latifolia (Halymeniaceae, Rhodophyta) from Europe with emphasis on cystocarp development and description of Nesoia mediterranea sp. nov. Phycologia 58, 393–404.
Rodríguez-Prieto, C., Afonso-Carrillo, J., De Clerck, O., Huisman, J. M., and Lin, S.-M. (2020). Systematic revision of the foliose Halymeniaceae (Halymeniales, Rhodophyta) from Europe, with the description of Halymenia ballesterosii sp. nov. from the Mediterranean sea and Nesoia hommersandii from the Canary Islands. Eur. J. Phycol. 55, 454–466.
Saunders, G. W., Chiovitti, A., and Kraft, G. T. (2002). Small-subunit rDNA sequences from representatives of selected families of the Gigartinales and Rhodymeniales (Rhodophyta). 3. Delineating the Gigartinales sensu stricto. Can. J. Bot. 82, 43–74. doi: 10.1139/b03-110
Saunders, G. W., and Kraft, G. T. (1996). Small-subunit rRNA gene sequences from representatives of selected families of the Gigartinales and Rhodymeniales (Rhodophyta). II. Recognition of the Halymeniales ord. nov. Can. J. Bot. 74, 694–707. doi: 10.1139/b96-088
Saunders, G. W., and Kraft, G. T. (2002). Two new Australian species of Predaea (Nemastomataceae, Rhdophyta) with taxonomic recommendation for an emended Nemastomatales and expanded Halymeniales. J. Phycol. 38, 1245–1260. doi: 10.1046/j.1529-8817.2002.02039.x
Schmitz, F. (1889). Systematische Übersicht der bisher bekannten Gattungen der Florideen. Flora Oder Algemeine Bot. Zeitung 72, 435–456.
Schneider, C. W., Popolizio, T. R., Kraft, L. G. K., and Saunders, G. W. (2019). New species of Galene and Howella gen. nov. (Halymeniaceae, Rhodophyta) from the mesophotic zone off Bermuda. Phycologia 58, 690–697. doi: 10.1080/00318884.2019.1661158
Schnetter, R., Richter, U., Schesmer, A., and Bula-Meyer, G. (1983). Licht- und Elektronenmikroskopische Untersuchungen an Grateloupiocolax colombiana gen. et spec. nov. (Halymeniaceae, Rhodophyceae). Beitr. Biol. Pflanzen. 58, 77–94.
Silva, P. C. (2002). Comments on the commentary by Kraft and Saunders [Phycologia 39, 258-261 (2000)] (Commentary). Phycologia 41, 99–100. doi: 10.2216/i0031-8884-41-1-99.1
Silva, P. C., and Johansen, H. W. (1986). A reappraisal of the order Corallinales (Rhodophyceae). Br. Phycol. J. 21, 245–254. doi: 10.1080/00071618600650281
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Wang, H. W., Kawaguchi, S., Horiguchi, T., and Masuda, M. (2001). A morphological and molecular assessment of the genus Prionitis J. Agardh (Halymeniaceae, Rhodophyta). Phycol. Res. 49, 251–262. doi: 10.1111/j.1440-1835.2001.tb00255.x
Wilkes, R. J., Lynne, M. M., and Guiary, M. D. (2005). Using rbcL sequence data to reassess the taxonomic position of some Grateloupia and Dermocorynus species (Halymeniaceae, Rhodophyta) from the north-eastern Atlantic. Eur. J. Phycol. 40, 53–60. doi: 10.1080/09670260400024634
Withall, R. D., and Saunders, G. W. (2006). Combining small and large subunit ribosomal DNA genes to resolve relationships among orders of the Rhodymeniophycidae (Rhodophyta), recognition of the Acrosymphytales ord. nov. and Sebdeniales ord. nov. Eur. J. Phycol. 41, 379–394. doi: 10.1080/09670260600914097
Wittmann, W. (1965). Aceto-iron-haematoxylin-chloral hydrate for chromosome staining. Stain Tech. 40, 161–164. doi: 10.3109/10520296509116398
Womersley, H. B. S., and Lewis, J. A. (1994). “Family Halymeniaceae Bory 1828, 158,” in The Marine Benthic Flora of Southern Australia. Part IIIA. Bangiophyceae and Florideophyceae (Acrochaetiales, Nemaliales, Gelidiales, Hildenbrandiales and Gigartinales sensu lato), ed. H. B. S. Womersley (Canberra: Australian Biological Resources Study), 167–218.
Yang, E. C., and Boo, S. M. (2004). Evidence for two independent lineages of Griffithsia (Ceramiaceae, Rhodophyta) based on plastid protein-coding psaA, psbA, and rbcL gene sequences. Mol. Phylo. Evol. 31, 680–688. doi: 10.1016/j.ympev.2003.08.014
Yang, E. C., Boo, S. M., Bhattacharya, D., Saunders, G. W., Knoll, A. H., Fredericq, S., et al. (2016). Divergence time estimates and the evolution of major lineages in the florideophyte red algae. Sci. Rep. 6:21361. doi: 10.1038/srep21361
Yang, E. C., Kim, K. M., Kim, S. Y., Lee, J.-M., Boo, G. H., Lee, J., et al. (2015). Highly conserved mitochondrial Genomes among multicellular red algae of the Florideophyceae. Genome Biol. Evol. 7, 2394–2406. doi: 10.1093/gbe/evv147
Yang, M. Y., and Kim, M. S. (2015). Taxonomy of Grateloupia (Halymeniales, Rhodophyta) by DNA barcode marker analysis and a description of Pachymeniopsis volvita sp. nov. J. Appl. Phycol. 27, 1373–1384. doi: 10.1007/s10811-014-0432-1
Yoon, H. S., Hackett, J. D., and Bhattacharya, D. (2002). A single origin of the peridinin- and fucoxanthin-containing plastids in dinoflagellates through tertiary endosymbiosis. Proc. Natl. Acad. Sci. U.S.A. 99, 11724–11729. doi: 10.1073/pnas.172234799
Keywords: female reproductive structures, Grateloupiaceae, Halymeniales, multigene phylogeny, taxonomy, tetrasporangial development
Citation: Kim SY, Lee HW, Yang EC, Boo SM, Lopez-Bautista J, Fredericq S, D’Archino R, Yoon HS and Kim MS (2021) Resurrection of the Family Grateloupiaceae Emend. (Halymeniales, Rhodophyta) Based on a Multigene Phylogeny and Comparative Reproductive Morphology. Front. Ecol. Evol. 9:775627. doi: 10.3389/fevo.2021.775627
Received: 14 September 2021; Accepted: 11 November 2021;
Published: 08 December 2021.
Edited by:
Joong-Ki Park, Ewha Womans University, South KoreaReviewed by:
Jeffery Hughey, Hartnell College, United StatesGwang Hoon Kim, Kongju National University, South Korea
Copyright © 2021 Kim, Lee, Yang, Boo, Lopez-Bautista, Fredericq, D’Archino, Yoon and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hwan Su Yoon, hsyoon2011@skku.edu; Myung Sook Kim, myungskim@jejunu.ac.kr
†These authors have contributed equally to this work
 Su Yeon Kim1,2†
Su Yeon Kim1,2†  Hyung Woo Lee
Hyung Woo Lee Eun Chan Yang
Eun Chan Yang Sung Min Boo
Sung Min Boo Suzanne Fredericq
Suzanne Fredericq Hwan Su Yoon
Hwan Su Yoon