Phylogeography of Blue Corals (Genus Heliopora) Across the Indo-West Pacific
- 1Interdisciplinary Graduate School of Agriculture and Engineering, University of Miyazaki, Miyazaki, Japan
- 2Department of Earth and Environmental Sciences (DISAT), University of Milano-Bicocca, Milan, Italy
- 3Marine Research and High Education Center (MaRHE Center), Faafu Magoodhoo, Maldives
- 4Department of Biological Sciences, National University of Singapore, Singapore, Singapore
- 5Tropical Marine Science Institute, National University of Singapore, Singapore, Singapore
- 6Centre for Nature-based Climate Solutions, National University of Singapore, Singapore, Singapore
- 7Division of Natural Sciences, University of Guam, Mangilao, GU, United States
- 8Coral Conservation and Research Group, Trace and Environmental DNA Laboratory, School of Molecular and Life Sciences, Curtin University, Bentley, WA, Australia
- 9Collections and Research, Western Australian Museum, Welshpool, WA, Australia
- 10Biodiversity Research Center, Academia Sinica, Taipei, Taiwan
- 11Parasitology, Faculty of Medicine, University of Miyazaki, Miyazaki, Japan
- 12Department of Life Science and Technology, School of Life Sciences and Technology, Tokyo Institute of Technology, Tokyo, Japan
- 13Okinawa Environment Science Center, Urasoe, Japan
- 14Biodiversity Program, Taiwan International Graduate Program, Academia Sinica and National Taiwan Normal University, Taipei, Taiwan
- 15Institute of Oceanography, National Taiwan University, Taipei, Taiwan
- 16Phuket Marine Biological Center, Muang, Thailand
- 17Faculty of Agriculture, University of Miyazaki, Miyazaki, Japan
Species delimitation of corals is one of the most challenging issues in coral reef ecology and conservation. Morphology can obscure evolutionary relationships, and molecular datasets are consistently revealing greater within-species diversity than currently understood. Most phylogenetic studies, however, have examined narrow geographic areas and phylogeographic expansion is required to obtain more robust interpretations of within- and among- species relationships. In the case of the blue coral Heliopora, there are currently two valid species (H. coerulea and H. hiberniana) as evidenced by integrated genetic and morphological analyses in northwestern Australia. There are also two distinct genetic lineages of H. coerulea in the Kuroshio Current region that are morphologically and reproductively different from each other. Sampling from all Heliopora spp. across the Indo-Pacific is essential to obtain a more complete picture of phylogeographic patterns. To examine phylogenetic relationships within the genus Heliopora, we applied Multiplexed inter simple sequence repeat (ISSR) Genotyping by sequencing (MIG-seq) on > 1287 colonies across the Indo-West Pacific. Maximum likelihood phylogenetic trees indicated the examined Heliopora samples comprise three genetically distinct groups: H. coerulea group, H. hiberniana group, and a new undescribed Heliopora sp. group with further subdivisions within each group. Geographic structuring is evident among the three species with H. hiberniana group found in the Indo-Malay Archipelago and biased toward the Indian Ocean whilst Heliopora sp. was only found in the Kuroshio Current region and Singapore, indicating that this taxon is distributed in the western Pacific and the Indo-Malay Archipelago. Heliopora coerulea has a wider distribution, being across the Indian Ocean and western Pacific. This study highlights the effectiveness of phylogenetic analysis using genome-wide markers and the importance of examining populations across their distribution range to understand localized genetic structure and speciation patterns of corals.
Introduction
Coral reef ecosystems have the highest biodiversity of all marine ecosystems (Fisher et al., 2015) and are important in terms of their scientific, economic, and social value (Moberg and Folke, 1999; Spalding et al., 2001; Cesar et al., 2003). However, reef-building corals, the foundation organisms in coral reefs, have been rapidly deteriorating (Hughes et al., 2003; Altieri et al., 2017; Eyre et al., 2018) due to coral mass bleaching and other threats associated with the on-going climate change (e.g., global warming and ocean acidification) (Hoegh-Guldberg et al., 2007; Hoegh-Guldberg and Bruno, 2010; Hoegh-Guldberg, 2011; Hughes et al., 2018); local anthropogenic impacts (e.g., red-soil runoff, nutrient enrichment, and marine plastic pollution) (Halpern et al., 2008; Omori, 2011; Huang et al., 2020; de Oliveira Soares et al., 2020); outbreaks of corallivorous animals (Birkeland and Lucas, 1990; Yasuda, 2018; Montalbetti et al., 2019) and coral disease (Bruno et al., 2003, 2007; Burge et al., 2014). Approximately one-third of the world’s reef-building corals are facing an elevated risk of extinction (Carpenter et al., 2008; Wilkinson, 2008), but many more cryptic species may be undescribed (Bickford et al., 2007) and if the natural range of species is misunderstood, their threatened status may be under-estimated (Richards et al., 2016).
The classification of reef-building coral species has traditionally been based mainly on morphological characteristics (Veron, 2000; Fabricius and Alderslade, 2001). In some cases, however, morphological classification has been difficult due to a high level of phenotypic plasticity across geographic and depth gradients, and in some genera, there is a lack of discrete morphological features that can be used to underpin species identifications (e.g., Forsman et al., 2009; Marti-Puig et al., 2014; McFadden et al., 2017). More recently, genetic information based on several molecular markers has shown morphology can conceal cryptic evolutionary relationships (Richards et al., 2013) and it is increasingly common for genetically distinct cryptic species or lineages to be found within widespread coral species (e.g., Nakajima et al., 2012; Pinzón et al., 2013; Warner et al., 2015). Such discordance between morphology and genetics has led to confusion regarding the species boundaries in many groups (e.g., Acropora, van Oppen et al., 2001; Marquez et al., 2002, Pocillopora, Flot et al., 2008; Souter, 2010; Schmidt-Roach et al., 2013). Such ambiguity threatens to undermine effective biodiversity conservation efforts and prevents the true complexity of coral reef ecosystems from being properly understood.
Molecular data is not, however, always able to provide a definitive answer about species boundaries within reef-building corals. Molecular systematic studies have been partly hindered by the lack of appropriate genetic markers and the traditionally used mitochondrial markers have suffered from inadequate analytical resolution (Shearer et al., 2002; Ridgway and Gates, 2006; Bilewitch and Degnan, 2011). The ability to interpret nuclear datasets is also hampered by the multi-copy nature of genes (Vollmer and Palumbi, 2004) even those that were originally thought to be single copy (e.g., Pax-C, Rosser et al., 2017), introgressive hybridization events (Veron, 1995; Richards et al., 2013) and/or incomplete lineage sorting (e.g., van Oppen et al., 2001; Souter, 2010; Yasuda et al., 2015). Recent advances in High-Throughput Sequencing (HTS) have enabled the use of genome-wide polymorphisms such as restriction site associated DNA sequencing (RAD-seq, Miller et al., 2007; Baird et al., 2008; Rowe et al., 2011; and Dart-seq, Rosser et al., 2017) and Multiplexed inter simple sequence repeat (ISSR) genotyping by sequencing (MIG-seq, Suyama and Matsuki, 2015) to infer phylogenetic relationships. Such techniques often successfully delimit species of non-model organisms including corals with higher resolution than traditional genetic markers (RAD-seq, e.g., Pante et al., 2015; Herrera and Shank, 2016; Quattrini et al., 2019 and MIG-seq, e.g., Tamaki et al., 2017; Park et al., 2019; Hirai, 2019).
MIG-seq is an easy, cost-effective, novel method to obtain a moderate number of single nucleotide polymorphisms (SNPs) of non-model organisms with polymerase chain reaction (PCR) and HTS technology. The number of SNPs from MIG-seq analysis is generally less than those obtained using other techniques such as RAD-seq. However, MIG-seq has several advantages, since the method can be performed with small amounts and/or low-quality DNA and is relatively easy and cheap. Indeed, MIG-seq successfully revealed species boundaries of octocoral species that could be undetectable by traditional genetic markers (Richards et al., 2018; Takata et al., 2019). While some studies examined genetic relationships of closely related reef-building corals using HTS techniques in geographically restricted regions (e.g., Forsman et al., 2017; Johnston et al., 2017), only a few studies have analyzed speciation and/or genetic divergence patterns covering the Indo-Pacific scale (but see Warner et al., 2015; Nakajima et al., 2017; Arrigoni et al., 2020; Wepfer et al., 2020). For a more comprehensive picture of the phylogeographic structure of coral species in the Indo-Pacific, and to understand the species diversity of corals, it is necessary to obtain more extensive representation of species across their geographic range.
Increasingly recognized is the importance of integrating information about reproductive timing and physiology with genetic and morphological data to obtain more robust estimates of coral species (see Ohki et al., 2015; Rosser, 2015; Villanueva, 2016; Luzon et al., 2017; Richards et al., 2018). Timing of reproduction in broadcast spawning species is particularly important because gametes are viable for only a few hours, so individuals that spawn more than a few hours apart are unlikely to cross-fertilize (Levitan et al., 2011) and in this regard, reproductive timing heavily influences evolutionary patterns. Whilst integrated datasets that include reproductive data and cover a wide geographic area are emerging as the gold standard in phylogenetic studies, practical and logistical constraints often mean it is not possible to obtain complete sampling coverage in every region (Keyse et al., 2014; Wepfer et al., 2021). Nevertheless, examining the species boundaries, ecological preferences, and distribution of reef-building corals is fundamental for promoting effective conservation and management strategies.
The blue corals (genus Heliopora) are members of octocorals, though it has a well-developed aragonite skeleton like those of scleractinians (Quattrini et al., 2020). Owning to their hard skeleton, Heliopora spp. play an important role in coral reef accretion in various Indo-West Pacific reefs (e.g., Zann and Bolton, 1985; Planck et al., 1988; Abe et al., 2008; Takino et al., 2010), and therefore is an important reef-building coral taxon. Heliopora coerulea (Pallas, 1766) has mainly three growth forms, namely lobate, columnar, and encrusting (Dana, 1846), but with high morphological plasticity and continuous variations among growth forms (Eguchi, 1948; Colgan, 1984; H. Taninaka pers. obs.). Heliopora coerulea has long been regarded as a living fossil, being the sole extant member of the family Helioporidae (Colgan, 1984) until the recent description of Heliopora hiberniana Richards et al., 2018, a morphologically, genetically and reproductively distinct species from northwestern Australia in the Indian Ocean (Richards et al., 2018). To date, H. coerulea is known to be distributed throughout the Indo-Pacific realm (Wells, 1954; Bouillon and Houvenaghel-Crévecoeur, 1970; Zann and Bolton, 1985), whereas H. hiberniana is known from the north-west shelf of Western Australia, the Maldives, and the Wakatobi and Gili Islands in Indonesia (Richards et al., 2018, 2020).
Heliopora spp. are gonochoric, brooding corals (Babcock, 1990) with short larval dispersal durations (Harii et al., 2002; Harii and Kayanne, 2003). Due to such a low larval dispersal ability, genetic connectivity among geographically separate populations may be limited, a pattern that was evident within the scale of several hundred square kilometers (Taninaka et al., 2019). Therefore, it is possible that Heliopora includes further genetically distinct lineages in the Indo-Pacific. Previous genetic studies have indicated that additional cryptic species may indeed be present within H. coerulea along the Kuroshio Current region of the Philippines, Taiwan and Japan (Yasuda et al., 2014). Study from Yasuda et al., 2014 hypothesized that there are at least two genetically distinct lineages (HC-A and HC-B) of H. coerulea in the Kuroshio Current region that can also be distinguished by different gross morphologies (small branch for HC-A and flat shapes for HC-B, Yasuda et al., 2014, 2015; Iguchi et al., 2019). Subsequent observations (Saito et al., 2015; Villanueva, 2016) and histological examination (Taninaka et al., 2018) revealed that the reproductive timing of the two lineages is different by almost one month even under similar environmental conditions. However, the phylogeographical relationships among HC-A and B in the Kuroshio Current regions, and with H. hiberniana and H. coerulea in northwestern Australia are still unknown. The aim of this study was to clarify the phylogenetic relationships and distributions of the genus Heliopora across the Indo-West Pacific to gain insights on the speciation patterns of Indo-Pacific coral species. To achieve this goal, we collected Heliopora samples widely from the Indo-West Pacific region and applied genome-wide phylogenetic, phylogeographic, and population genetic analyses using the MIG-seq method.
Materials and Methods
Sample Collection and DNA Extraction
A total of 1287 Heliopora samples from a total of 73 sites across three western Pacific regions (Japan, Taiwan and Guam) and Singapore, and three Indian Ocean regions (northwestern Australia, Thailand, and the Maldives) were collected using SCUBA from 2008 to 2019 (Supplementary Table 1). Additionally, we used previously collected samples from Japan and northwestern Australia (Supplementary Table 1). The samples include those from the northernmost distribution site (Yaku Island in Japan; Nakabayashi et al., 2017), and the known deepest depth (from 50 m depth from Green Island, Taiwan). All coral fragments were preserved in 99.5% EtOH and kept at −20°C until DNA extraction. During sampling, colonies were photographed, and GPS information was acquired wherever possible. All overseas sample collection was conducted through legal procedures and collection cooperation with the support of local research collaborating institutions; Academia Sinica in Taiwan, University of Guam in Guam, National University of Singapore in Singapore, Western Australian Museum and Curtin University in Western Australia, Phuket Marine Biological Center in Thailand, University of Milano-Bicocca and Marine Research and High Education Center in Maldives (see also FIELD STUDY PERMISSION). Genomic DNA was extracted from the preserved samples using the hot alkaline solution method (Meeker et al., 2007) or QIAGEN DNeasy Blood & Tissue kit (QIAGEN, Hilden, Germany) on site. The first PCR procedure of MIG-seq analysis was carried out at each overseas laboratory except for Thailand samples from which ethanol preserved coral fragments were imported in 2011 (CITES number: AC.0510.2/407). We used them in this study under an additional permit from the Department of Marine and Coastal Resources in Thailand.
MIG-seq Library Preparation and Sequencing
All the DNA extraction and first PCR procedure except for domestic samples were carried out locally, and the first PCR products were transported to Miyazaki, Japan. The first PCR step was performed by using 8 pairs of multiplex ISSR primers of Suyama and Matsuki (2015). The fragments were amplified with the Multiplex PCR Assay Kit Ver.2 using a total volume of 7 μL reaction in a thermal cycler with the following modified profile: 94°C for 1 min followed by 29 cycles at 94°C for 30 s, 38°C for 1 min, 72°C for 1 min, and a final extension at 72°C for 10 min. The 50-fold diluted first PCR product was used as the template DNA for the second, tailed-PCR to add the individual index and the Illumina adapter sequence to each sample. PrimeSTAR GXL DNA polymerase (TaKaRa Bio Inc., Otsu, Shiga, Japan) was used for second PCR with a total volume of 12 μL reaction in a thermal cycler with a following profile: 20 cycles at 98°C for 10 s, 54°C for 15 s, 68°C for 1 min. The second PCR products were pooled as a single mixture library by using 1μL of each product. The mixed sample was electrophoresed on 0.1% agarose gel to verify amplification. The size ranged from 350 to 800 bp and DNA was manually extracted from the gel on a transilluminator. After extracting genomic DNA from the gel using FastGene Gel/PCR Extraction Kit (Nippon Genetics, Tokyo, Japan), library concentration was measured with a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, United States). Finally, the DNA library was sequenced on a MiSeq Sequencer (sequencing control software v2.0.12, Illumina, San Diego, CA, United States) using a MiSeq Reagent Kit v3 (150 cycles; Illumina). Image analysis and base calling were performed using real-time analysis software v1.17.21 (Illumina). DarkCycle option was changed “Amplicon-dark 17-3” to “Amplicon-dark 17-17” on the “Chemistry” line (see also Suyama and Matsuki, 2015).
Sequence Processing and SNP Genotyping
The FASTX-toolkit version 0.0.14 (Gordon and Hannon, 2012), with a fastq-quality-filter setting of –Q 33 –q 30 –p 40, was used to eliminate low-quality reads and primer sequences from the MIG-seq raw data. Adapter sequences for Illmina MiSeq were removed from both 5’ end and 3’ end using Cutadapt version 2.10 (Martin, 2011). Short reads less than 80 bp were excluded using an in-house python script. To further exclude contamination from Symbiodiniaceae-DNA, and to obtain larger numbers of loci (Shafer et al., 2017), the filtered raw reads were mapped onto the reference genome of Heliopora coerulea obtained from Symbiodiniaceae-free larvae (10.6084/m9.figshare.14356418) using Stacks version 2.2.0 (Catchen et al., 2011; Rochette et al., 2019) with the reference-aligned pipeline for single nucleotide polymorphism (SNP) discovery and genotyping. As a first step, the unpaired reads were removed using the repair.sh software tool within BBtools software package (Bushnell, 2017). Next, the retained reads were aligned to the reference genome using Burrows-Wheeler Aligner (BWA) version 0.0.17-r118 (bwa index and mem with default parameters) (Li and Durbin, 2009). Alignments were then compressed, sorted, and indexed with SAMtools version 1.10 (Li et al., 2009). Finally, SNP genotyping for each individual was carried out using the gstacks program in Stacks with default setting. SNP calling for each genetic analysis was carried out using the populations program in Stacks with the option -r to obtain different minimum proportions of genotyped individuals per locus (0.1 for phylogenetic analyses, 0.9 for assigning clones and population genetic analyses), and the option –ordered_export to ensure that only one of the overlapping SNPs is output from reference aligned data. All the output files from Stacks were converted to the appropriate formats for each genetic analysis using PGDSpider version 2.1.1.5 (Lischer and Excoffier, 2012).
Assign Clones
We used the software GenoDive version 3.04 (Meirmans, 2020) to assign possible clones within each population based on the infinite allele model (IAM) omitting all missing data. Less than 12 differences of multi-locus genotype within the same population were identified as possible clone mates (estimated by inspection of the pairwise distance histogram in GenoDive). All clone mates except for one individual with the least missing data were removed from this study. After removing possible clones, we selected up to three least missing data samples per sampling site. In case multiple different gross morphologies and/or genetically different lineages were found within the same site, up to three samples per morphology were selected. Finally, we rerun Stacks using this selected dataset excluding possible clones to obtain new SNPs for phylogenetic and population genetic analyses.
Phylogenetic Analysis
We estimated phylogenetic trees based on the Maximum likelihood (ML) method. The program IQ-TREE2 version 2.0.6 (Minh et al., 2020), using the TVM + F + R5 model selected based on the Bayesian Information Criterion (BIC), was used to reconstruct a phylogenetic hypothesis for Heliopora spp. The bootstrap analysis was performed with 1000 replicates using UFBoot, Ultrafast Bootstrap Approximation (Minh et al., 2013). The final tree was drawn using FigTree version 1.4.4 (Rambaut, 2012).
Population Genetic Analysis
We conducted Principal Coordinates Analysis (PCoA) to examine genetic relationships among Heliopora individuals using GenAlEx version 6.502 (Peakall and Smouse, 2012). In addition, Discriminant Analysis of Principal Components (DAPC) (adegenet package version 2.1.3 in R version 4.0.2) (Jombart, 2008; Jombart et al., 2010; Jombart and Ahmed, 2011) was performed to visualize the genetic structure. The lowest BIC was used to detect the optimal number of K clusters.
Results
Number of Reads and SNPs From MIG-seq Analysis
In total 339,818,358 raw reads with an average of 264,039 reads per sample were obtained for 1,287 individuals. After filtering low-quality reads, 335,164,915 reads with an average of 260,423 reads per sample were obtained. After excluding index, adapter, and short reads less than 80 bp, 139,837,592 reads with an average of 108,654 reads were obtained. In total 67,119,478 reads were mapped onto the reference-genome with an average of 52,152 reads per sample. We used 795 SNPs across 1,287 individuals (r = 0.9) to find possible clones, and then excluded 576 individuals from the subsequently genetic analyses. We finally selected 245 individuals from 73 sampling sites from 7 regions (DRA Accession No. DRA012077) for phylogenetic (ML tree; 24,741 SNPs, r = 0.1) and population genetic analyses (PCoA and DAPC; 1,092 SNPs, r = 0.9). For the latter, we selected 238 individuals from the final dataset after excluding seven individuals with a missing rate of more than 50% per 1,092 SNPs.
Phylogenetic Relationships
ML phylogenetic analysis revealed three major groups of Heliopora spp. in the Indo-West Pacific with 100% bootstrap values (Figure 1). The phylogenetic hypothesis presented here indicated that there is one new undescribed group (hereafter Heliopora sp. group) genetically different from the other two groups, each of which includes typical morphologies of the previously described species (slender-branching growth form H. hiberniana whose type locality is in the northwestern Australia; lobate growth form H. coerulea whose type locality is in the Indian Ocean). The “Heliopora sp.” group (shown in blue in Figure 1) comprises the HC-A lineage found in Kuroshio Current regions including Japan and Taiwan (Yasuda et al., 2010, 2014, 2015; Taninaka et al., 2019), and Singapore. The “H. hiberniana” group (shown in green in Figure 1) includes the holotype of H. hiberniana found in northwestern Australia (Richards et al., 2018) and the eastern Indian Ocean samples from Thailand and the Maldives. The “H. coerulea” group (shown in red in Figure 1) comprises mainly lobate growth morphs HC-B lineage found in Kuroshio Current regions including Japan and Taiwan (Yasuda et al., 2010, 2014, 2015; Taninaka et al., 2019), Guam, and H. coerulea found in northwestern Australia (Richards et al., 2018). In addition, there were genetically isolated subclades within each group that clustered together by different growth forms or geographic regions (Figure 1; sp.1–3, hib.1–2, and coe.1–3). The Heliopora sp. group was divided into three subclades: Singapore subclade (sp.1), HC-A with only columnar growth form subclade of Japan (sp.2), and HC-A with both encrusting and columnar growth forms subclade found in both Japan and Taiwan (sp.3). Heliopora sp.3 also included a sample collected from 50m depth in Green Island, Taiwan. A sample collected from Yaku Island in Japan, the northernmost distribution site, looks like an outgroup of sp.2 and sp.3 in the phylogeny, possibly because it had a hybrid genotype between Heliopora sp.2 and sp.3 based on STRUCTURE analysis (data shown in Supplementary Figure 1). The group “H. hiberniana” was divided into two subclades: northwestern Australia H. hiberniana subclade (hib.1) and a subclade consisting of Thailand and the Maldives samples (hib.2). The group “H. coerulea” was divided into three subclades; a Guam subclade (coe.1), H. coerulea in northwestern Australia subclade (coe.2), and a subclade consisting of HC-B found in Japan and Taiwan samples (coe.3).
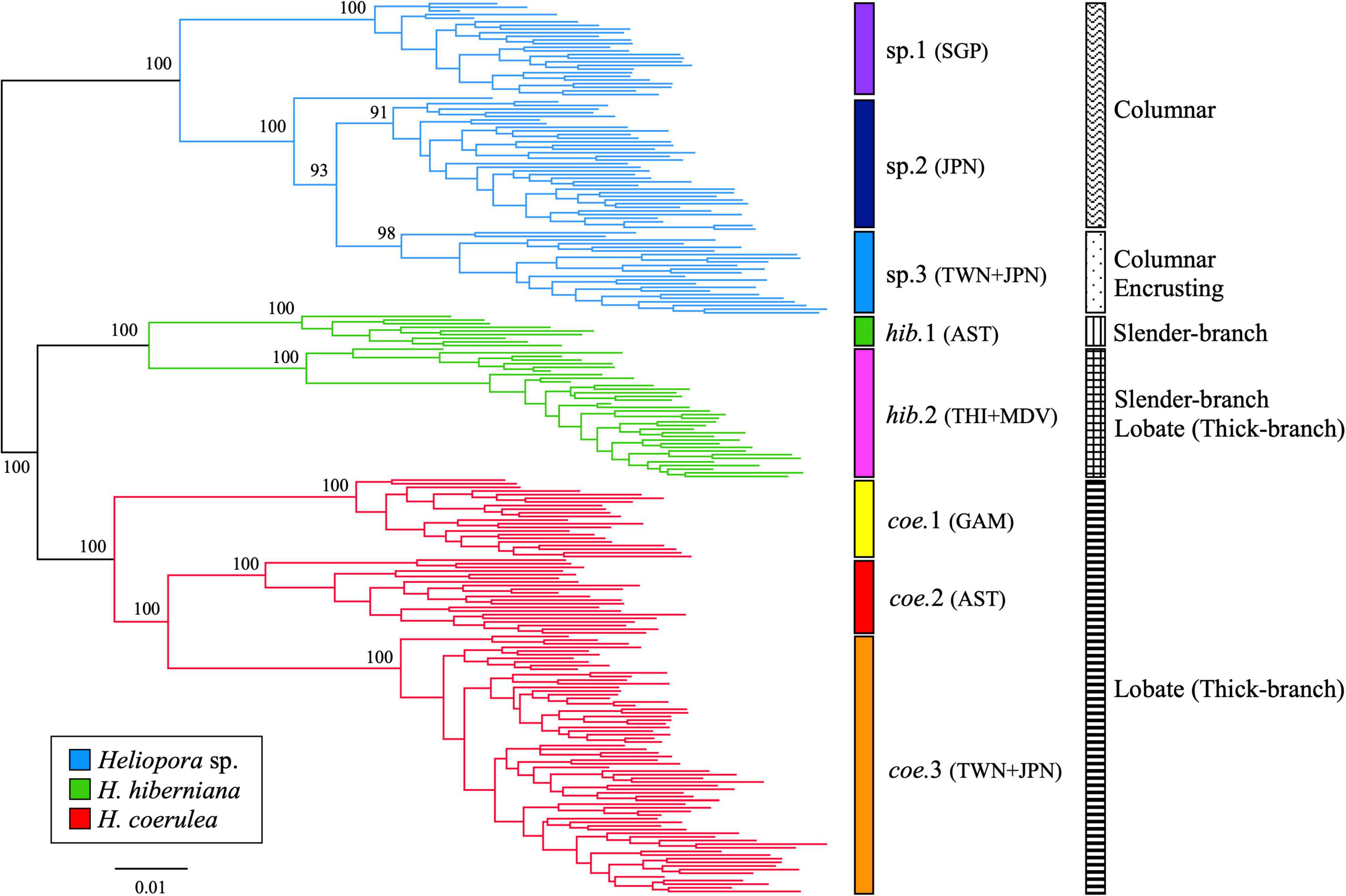
Figure 1. Maximum likelihood phylogeny of Heliopora spp. Node supports are provided if bootstrap values ≧90%. Blue Heliopora sp. group composed of three subclades; Singapore subclade (sp.1), a subclade of HC-A with columnar growth form found in Japan (sp.2), and a subclade of HC-A with both encrusting and columnar growth forms found in Japan and Taiwan (sp.3). Green H. hiberniana group composed of two subclades; a subclade including the holotype of H. hiberniana found in northwestern Australia (hib.1) and a Thailand and the Maldives subclade (hib.2). Red H. coerulea group composed of three subclades; Guam subclade (coe.1), a subclade of H. coerulea in northwestern Australia (coe.2), and a subclade of HC-B found in Japan and Taiwan (coe.3).
Individual-Based Genetic Differentiation
Consistent with the phylogenetic analysis result, population genetic analyses revealed three genetic isolated groups of Heliopora with eight subclades in the Indo-West Pacific (Figures 2A,B). The PCoA showed three clusters based on 11.73% (PC1) and 7.3% (PC2) of the total variability (Figure 2A). The DAPC also showed clear genetic separation of the three clusters and eight subclades (Figure 2B). K = 3 was selected supporting the ML phylogeny result, while K = 8 was determined as the optimal number of clusters by calculating the first 200 PCs for the maximum of 50 clusters based on BIC. The posterior DAPC assignments of both K = 3 and 8 were consistent with the prior clusters (all blue crosses were on red rectangles).
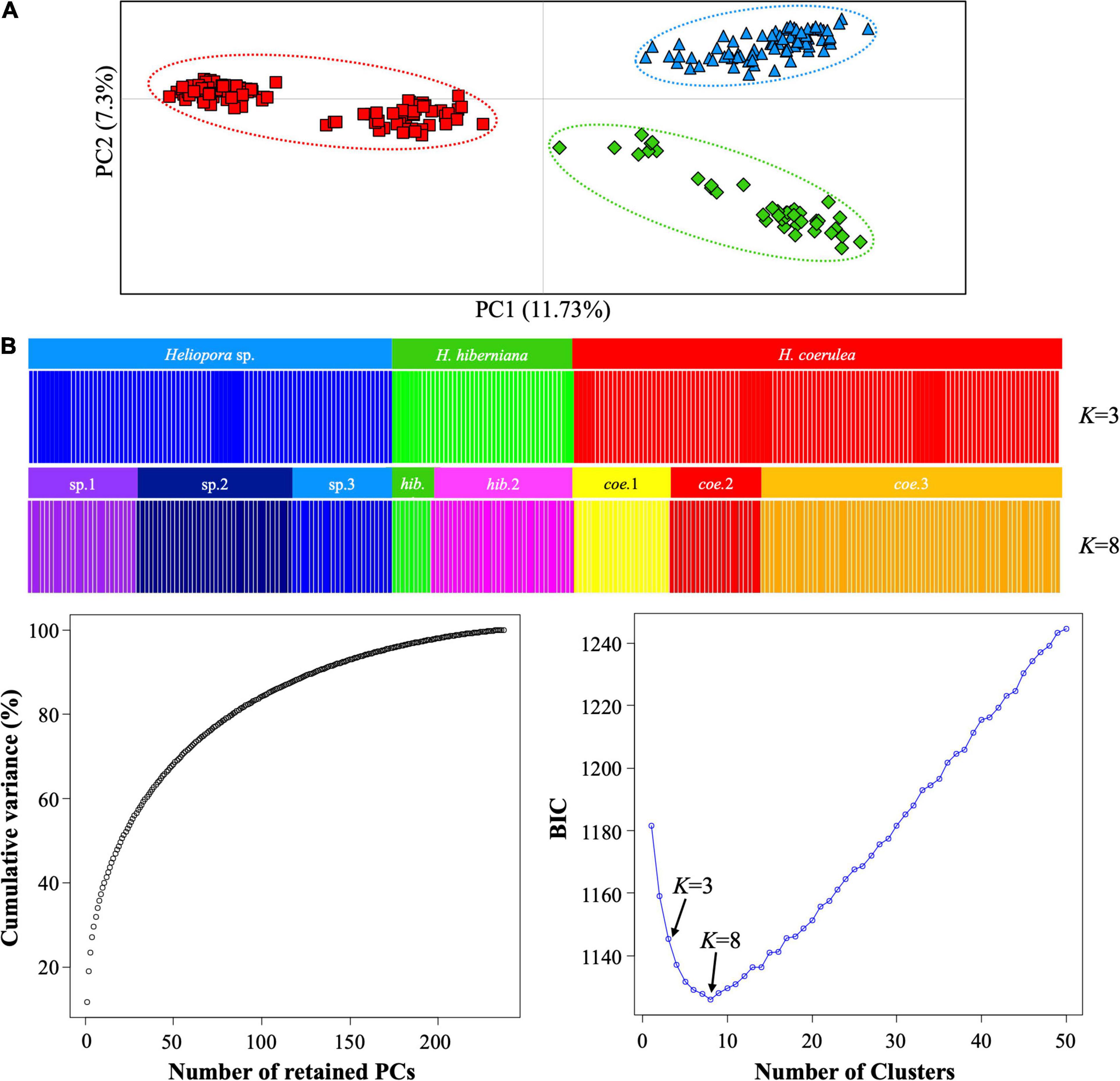
Figure 2. (A) Principal coordinates analysis calculated by GenAlEx v.6.502 using pairwise codominant genotypic distance. Each point represents an individual. (B) Genotype clusters composition plots (above two), and a graph of inference of the number of the PCs and BIC values (below two) of the DAPC. Each bar represents an individual and the colors represent proportional membership coefficients, K = 3 based on 2 PCs and K = 8 based on 7 PCs groups. All the colors used in (A) and (B) correspond with those in the ML tree (Figure 1).
Geographical Distribution
The three groups of Heliopora indicated uneven geographical distribution (Figure 3A). The distribution of Heliopora sp. group was biased to western Pacific Ocean side including Kuroshio Current region (Japan and Taiwan) and the South China Sea (Singapore). On the contrary, the distribution of H. hiberniana group was mainly in the Indian Ocean side such as Kimberley region and Christmas Island in northwestern Australia, the Andaman Sea (Thailand), and the Maldives. The H. coerulea group was distributed both in the Indian Ocean and western Pacific ranging from the Guam and the Kuroshio Current region (Japan and Taiwan) to Kimberley and Christmas Island in northwestern Australia. The distribution of H. coerulea group was partly overlapping with the other two species groups in Japan, Taiwan, and northwestern Australia (Figure 3A, coe.2 and hib.1; coe.3 and sp.2-3).
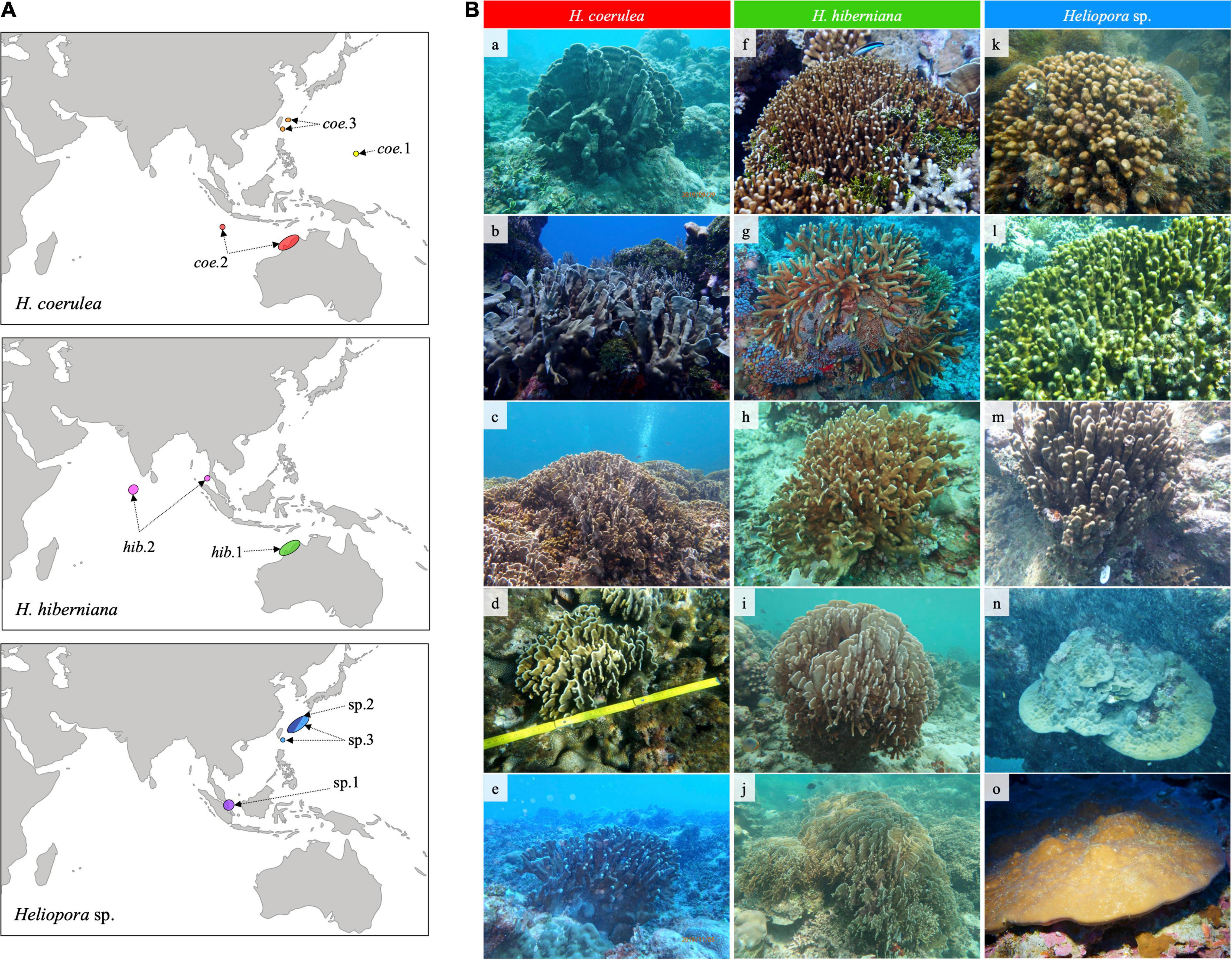
Figure 3. (A) Geographical distributions of the three Heliopora spp. groups. (B) Gross morphologies of the three Heliopora spp. groups. H. coerulea group; (a) lobate growth form in Guam (coe.1), (b) lobate growth form in northwestern Australia (coe.2), (c) lobate growth form in Japan and (d) Taiwan (coe.3), and (e) thick-branching growth in Guam (coe.1). H. hiberniana group; (f) slender-branching form in the type locality in northwestern Australia (hib.1), and (g) slender-branching and (h–j) lobate growth form in the Maldives (hib.2). Heliopora sp. group; (k) columnar growth form in Singapore (sp.1), (l) columnar growth form in Japan (sp.2), (m) columnar growth form in Japan (sp.3), (n) encrusting growth form in Japan (sp.3), and (o) encrusting growth form in the mesophotic zone (50 m) in Taiwan (sp.3).
Gross Morphology
The gross morphologies of the three groups of Heliopora were highly variable (Figure 3B). H. coerulea group included two typical morphs; lobate growth form (Figure 3B, a–d) and thick-branching growth form (Figure 3B, e). H. hiberniana group included two typical morphs; slender-branching growth form (Figure 3B, f,g) and lobate growth form (Figure 3B, h–j). Heliopora sp. group included two typical morphs; columnar growth form (Figure 3B, k–m) and encrusting growth form (Figure 3B, n,o). The typical growth forms of Heliopora sp. group were relatively distinct from other two species, while the morphologies of H. hiberniana group and H. coerulea group partly overlapped and were difficult to identify by gross morphology.
Discussion
In the present study, we examined the phylogenetic relationships, gross morphological variations, and the geographical distributions of Heliopora spp. collected from the Indo-West Pacific region. Our phylogenetic reconstruction indicates that there are three genetically isolated species groups in the genus Heliopora and we provide evidence that there is substantial subcladal genetic structuring within each group.
We do not describe the newly found Heliopora sp. group or its subclades as new species of the genus Heliopora in this study. Cladistic analysis and formal species description are being undertaken separately.
Phylogeography and Speciation Patterns of Heliopora
Phylogenetic and population genetic analyses using genome-wide SNPs by MIG-seq showed that the Indo-West Pacific Heliopora was divided into three groups: H. coerulea group, distributed in the Indo-West Pacific and including HC-B in the Kuroshio Current region; H. hiberniana group distributed mainly in the Indian Ocean including the type locality of H. hiberniana in northwestern Australia; and a new undescribed Heliopora sp. group distributed mainly in the Western Pacific Ocean, including HC-A in the Kuroshio Current region (Figures 1, 2 and 3A; Supplementary Table 1).
The new undescribed Heliopora sp. group was distributed mainly in the Western Pacific (Japan and Taiwan) and Singapore in this study, while H. hiberniana group was distributed mainly in the Indian Ocean (northwestern Australia, the Andaman Sea in Thailand, and the Maldives in this study; distributed also in Indonesia, Richards et al., 2020). Such uneven geographical distributions imply the origin of Heliopora sp. group could be in the Pacific Ocean and that of H. hiberniana could be in the Indian Ocean, but this remains to be tested with a dated phylogeny. Cenozoic tectonic events (Hall and Holloway, 1998) and sea-level fluctuations associated with climate change (Pillans et al., 1998) in the area between the Indian and Pacific Oceans are thought to have promoted allopatric speciation in many coral reef organisms (Carpenter et al., 2011) including starfish (Benzie, 1999; Vogler et al., 2008) and reef fishes (Gaither et al., 2011; Bowen et al., 2013). It is, therefore, possible that the genus Heliopora has also allopatrically speciated into the two groups, Heliopora sp. and H. hiberniana, due to the physical division between the Indian and Pacific Oceans caused by past tectonic and climatic changes. Morphological and molecular analyses on other Indo-Pacific reef-building coral species also found sibling species between the Indian and Pacific Oceans (Veron, 1995; van Oppen et al., 2001; Pinzón et al., 2013; Huang et al., 2014; Arrigoni et al., 2020; Wepfer et al., 2020).
The H. coerulea group is distributed in both the Indian and Western Pacific Oceans. The distribution of H. coerulea group partially overlaps with the Heliopora sp. group in the western Pacific and with the H. hiberniana group in northwestern Australia. Previous studies revealed sympatrically distributed Heliopora spp. have different reproductive timing: the reproductive timing of Heliopora sp. group (HC-A) and H. coerulea group (HC-B) in Japan (Saito et al., 2015: Taninaka et al., 2018) and the Philippines (Villanueva, 2016) differ by almost one month. Heliopora hiberniana and H. coerulea colonies in northwestern Australia also have been found to have different reproductive timings even when found in sympatry (Richards et al., 2018). These patterns suggest that endogenetic regulation may contribute to differences in reproductive timing. A previous study revealed that there are fixed species-specific substitutions in the dopamine receptor 2-like gene (Isomura et al., 2013) and cryptochrome-1 (Oldach et al., 2017), both of which were identified as the genes that regulate reproductive timing in Acropora species (Iguchi et al., 2019). It is possible that allochronic speciation have occurred between these sympatrically distributed Heliopora spp. groups, or at least allochronism in reproductive timing plays a central role to keep their species boundaries after speciation. The difference in reproductive timing is an important mechanism as a prezygotic isolation for marine species that release gametes into the water column (Knowlton, 1993; Palumbi, 1994). Indeed, many of the sympatrically distributed broadcast spawning coral species have different reproductive timing in the same genus (e.g., Orbicella, Knowlton et al., 1997; Szmant et al., 1997; Acropora, Fukami et al., 2003; Nakajima et al., 2012; Ohki et al., 2015). Generally, the reproductive timing of corals is also strongly associated with environmental factors (e.g., moon phases, moonlight, solar radiation, water temperature) in addition to genetic factors (e.g., circadian clock genes, photoreceptor proteins) that correlate with the environmental factors (e.g., Levy et al., 2007; Brady et al., 2009; Crowder et al., 2017; Oldach et al., 2017). Thus, future studies on reproductive timing of Heliopora spp. that include both environmental and genetic factors may provide further insights into speciation of sympatrically distributed Heliopora spp.
The Heliopora sp. group was found both in Yaku Island in Japan (N 30°16′21.45″, Nakabayashi et al., 2017), the northernmost site and at a depth of 50 m on Green Island in Taiwan, the deepest habitat records of the genus Heliopora. Therefore, contrary to previous knowledge on the ecological traits of H. coerulea that it prefers warm shallow water habitats (Zann and Bolton, 1985), Heliopora sp. group could be found in colder and deeper habitat; the northernmost known habitat of H. coerulea group (HC-B) is Miyako Island (N 24°51′52.10″, Yasuda et al., 2014) and that of Heliopora sp. group (HC-A) is Yaku Island (N 30°16′21.45″, Nakabayashi et al., 2017). In the field, contrasting ecological differences have been previously observed for Heliopora sp. group (HC-A) and H. coerulea group (HC-B). Colonies of Heliopora sp. group (HC-A) are relatively small and more abundant outside the well-developed fringing reef, where water temperature is lower, while colonies of H. coerulea group (HC-B) can be larger, forming micro-atolls, and dominating inner reef habitats where water temperature is higher (Yasuda et al., 2010; Taninaka et al., 2018), providing evidence of niche differentiation which further support the hypothesis of ecological speciation between these two groups.
Gross Morphological Diversity of Heliopora
There are some specific growth forms within each Heliopora group. Columnar and encrusting growth forms are relatively distinct from other morphologies and specific to Heliopora sp. group (Figure 3B, k–o). Slender-branching growth form with whitish skeleton (Figure 3B, f–g) is also specific to H. hiberniana group. However, lobate growth form (Figure 3B, a–d and h–j) can be found in both H. coerulea and H. hiberniana groups. Like other reef-building coral species, young and/or juvenile stages are hard to distinguish among closely related species (Babcock, 1992; Babcock et al., 2003). Previous studies have also reported intermediate growth forms sometimes rendering it difficult to distinguish even between HC-A (Heliopora sp. group) and HC-B (H. coerulea group) possibly due to infrequent hybridization and phenotypic plasticity in the field (Yasuda et al., 2014; Taninaka et al., 2018). Therefore, not all colonies can be clearly classified into the Heliopora groups based on field observation alone.
Phenotypic plasticity and continuous morphological variation among closely related coral species are a common phenomenon and have hindered accurate morphological species identification (e.g., Forsman et al., 2009; Marti-Puig et al., 2014; McFadden et al., 2017). Light intensity and water movement are the most influential environmental factors responsible for coral phenotypic plasticity (Todd, 2008), while other physical environmental factors (e.g., dredging, sediment disturbances, turbidity, competition) are also correlated with morphological diversity, leading to continuous morphological variation of different species under similar environmental conditions (e.g., Darling et al., 2012; Erftemeijer et al., 2012; Swierts and Vermeij, 2016). However, it is still unclear what environmental factors influence gross morphological diversity in Heliopora spp.
Richards et al. (2018) previously reported the key morphological characteristics to distinguish H. coerulea from H. hiberniana, including gross morphology, the intensity of bluish skeletal color, and skeletal microstructure (e.g., highly elaborated coenchymal echinulations and smaller and more numerous autopores), indicating skeletal color can be a key morphological characteristic. In the western Pacific, a few colonies with whitish skeletons have been found in the Heliopora sp. group in Japan (H. Taninaka and N. Yasuda pers. obs.). Contrary to the case in the northwestern Australian region, there was no genetic difference between the whitish and other bluish skeleton colonies, indicating skeletal color cannot be a universal key morphological character to distinguish Heliopora sp. from the other species.
Based on these findings, it is necessary to clarify the relationship between morphological diversity including fine skeletal structure and environmental and/or genetic factors among Heliopora spp. in more detail in the future.
Subclades Within Each Heliopora Group
Our phylogenetic tree showed substantial phylogenetic structure and multiple genetically isolated subclades exist within each Heliopora species group. These subclades partially correspond to typical growth forms and geographical distributions (Figures 1, 2 and 3A). In the Heliopora sp. group for example, the geographically distant Singapore subclade (sp.1) first diverged from Japan-Taiwan clades (sp.2 and 3). Then, the Japan-Taiwan clades were split into only columnar growth form subclade (sp.2) found only in Japan and encrusting and columnar growth forms subclade (sp.3) found in both Japan and Taiwan. These two subclades were genetically distinct despite being sympatric. In the field, some ecological differences can be observed between the two subclades: Heliopora sp.2 tends to be found in shallow inner reef areas with strong sunlight and relatively weak waves. Heliopora sp.3 tends to be found in relatively deep darker areas such as outer reef slope with fast water currents and turbidity (H. Taninaka pers. obs.). Further ecological and physiological differences between Heliopora sp.2 and Heliopora sp.3 would clarify the species status of the two subclades. The Heliopora sp.2 and Heliopora sp.3 subclades could not be distinguished in the previous studies using traditional genetic markers (e.g., mitochondrial DNA, ITS2, microsatellite and microsatellite flaking region sequences) (Yasuda et al., 2014, 2015; Taninaka et al., 2019), while this study corroborated the effectiveness of using genome-wide SNPs to elucidate detailed genetic boundaries between closely related lineages/species as in the case of other octocorals (Herrera and Shank, 2016; Quattrini et al., 2019; Takata et al., 2019).
In the H. hiberniana group, two subclades (hib.1-2) showed deep genetic divergence corresponding to geographical distance, namely the northwestern Australian subclade (hib.1) and the Maldives-Thailand subclade (hib.2). This genetic isolation could be the result of long-term restriction of gene flow between the two subclades. The current circulation system in the Indian Ocean mainly consists of currents that move from east to west that does not directly connect northern (Maldives and Thailand) and southern (northwestern Australia) sides of the mid-east Indian Ocean (Schott and McCreary, 2001; Schott et al., 2009). Due to such current patterns, previous studies on other marine organisms with pelagic larval phase also showed similar genetic isolation in the Indian Ocean (Benzie et al., 2002; Bay et al., 2004; Hui et al., 2016). On the other hand, geological research (Schettino and Turco, 2011; Keith et al., 2013; Obura, 2016), fossils, and phylogenetic data (Obura, 2012, 2016; Veron et al., 2015) propose a hypothesis that tectonic changes occurred during the Paleogene and the Neogene and may have promoted genetic isolation and speciation events in the Indian Ocean. Such historical geographic events might have promoted genetic divergence of Heliopora spp. in the Indian Ocean, but this requires further testing within a calibrated phylogenetic framework.
In the H. coerulea group, three subclades (coe.1–3) corresponding to geographically distinct regions were found, namely Guam subclade (coe.1), the northwestern Australia subclade (coe.2), and Japan-Taiwan subclade (coe.3). The most genetically isolated subclade that first split from the others is the Guam subclade (coe.1). It is notable that Japan-Taiwan subclade (coe.3) is genetically more closely related to the northwestern Australia subclade (coe.2) in the Indian Ocean than to the geographically closer Guam subclade (coe.1). It is likely that genetic connectivity has been restricted between Guam and Japan-Taiwan since the Guam subclade (coe.1) diverged from the other two subclades early in the history of H. coerulea. This is unusual because genetic connectivity between Mariana Islands region including Guam and the Kuroshio Current region is often strong in most of the marine invertebrate species with larval dispersal period (e.g., Palumbi et al., 1997; Williams and Benzie, 1998; Lessios et al., 2001; Pinzón et al., 2013; Arrigoni et al., 2020; Wepfer et al., 2020; but see exception in Wörheide et al., 2008). Although the reasons for the genetic break between Guam and Kuroshio Current region in Heliopora spp. are unclear, it could be caused by limited larval dispersal capability of Heliopora spp. (Taninaka et al., 2019) and/or local adaptation of each subclade. On the other hand, northwestern Australian populations of other marine species are genetically more similar to west Pacific populations than other Indian Ocean populations (e.g., Williams and Benzie, 1998; Lessios et al., 2001; Pinzón et al., 2013; Arrigoni et al., 2020; Wepfer et al., 2020) but contrary examples also exist where Indian and Pacific Ocean coral populations are divergent (e.g., Richards et al., 2016). Williams and Benzie (1998) discussed that this might be due to Western Australia being recently colonized by recruits from the Western Pacific lineage after the end of the last ice age. Recent genome-wide phylogeographic analysis of reef-building corals have also found such large discrepancies between geographic and genetic distances (Arrigoni et al., 2020; Wepfer et al., 2020), which need to be examined in closer detail in the future.
Conclusion
The present study provides a comprehensive picture of phylogenetic relationships, distribution, and growth form patterns among closely related Heliopora species that gives new insights into speciation patterns of Heliopora spp. in the Indo-West Pacific. Our genome-wide genetic data of Heliopora spp. revealed three major groups with eight subclades. Heliopora sp. group was genetically distinct from the other two groups (H. coerulea group and H. hiberniana group) containing the samples from type localities of the previously described species. Genetic diversification in the genus Heliopora could be attributed to allopatry, allochrony, and local and/or ecological adaptation. Further studies incorporating more genome-wide markers for phylogeographic analysis, geohistorical information, physiological and ecological data would more precisely delineate the boundaries of Heliopora species.
Our study also demonstrates the effectiveness of the MIG-seq method for clarifying the species boundaries of octocoral species that were indistinguishable using traditional genetic markers such as mtDNA and ITS2. In addition, we highlight the importance of wide geographic sampling to generate a more complete picture of the phylogeographic relationships and speciation patterns among closely related octocoral species and how they diversified across the Indo-Pacific. It is expected that the MIG-seq method can be applied to other coral species and non-model organisms to resolve longstanding questions in marine evolution.
Field Study Permission
Domestic sampling except for Amami Oshima was conducted under each prefectural government permits; Yaku Island (Kagoshima Prefectural Government permit, No. 2–56), Okinawa including Okinawa Mainland, Kume Island, Ishigaki Island, Sekisei Lagoon, and Iriomote Island (Okinawa Prefectural Government permits, Nos. 18–34, 19–39, 19–60, 21–18, 22–15, 25–44, 26–10, 27–29, 27–81, 28–78, 30–42, and 31–18), and Amami Oshima was conducted under personal permit by Mr. Katsuki Oki. Foreign sampling was conducted under each local government permits; Taiwan (samples collected from Dabaisha and Gongguan, the Taitung County Government, No. 1074150258; a 50 m specimen collected in Chaiko, Taitung County Government, No. 1040000285, and that is registered at the Zoological collection of the Biodiversity Research Museum in Academia Sinica AZIC0001313), Guam (SC-19-006 (occ), University of Guam Marine Laboratory), Singapore (National Parks Board Permit NP/RP16-156), Australia (samples in Western Australia were collected under permit number AU-COM2013-203 and Australian Government approval number 006-RRRW-130723-01), Thailand (the permission for collecting and sending the specimens is granted by the Department of Fisheries), Maldives (Permit No. (OTHR)30-D/INDIV/2017/123 released by the Maldivian Ministry of Fisheries and Agriculture). All the 1st PCR except for those from Thailand were conducted on site and only 1st PCR products were brought or sent to Japan. Ethanol-preserved coral fragment samples from Thailand were imported to Japan in 2011 (CITES number: AC.0510.2/407), and we used them in this study under an additional permit from the Department of Marine and Coastal Resources in Thailand.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: DDBJ (accession: DRA012077), FigShare (doi: 10.6084/m9.figshare.14356418 and doi: 10.6084/m9.figshare.14814231).
Author Contributions
HT and NY conceived the study and drafted the manuscript. All authors except for TK and HY collected samples and conducted molecular genetic experiment. HT, TK, and HY conducted genetic analysis. All authors edited the draft and approved it for publication.
Funding
Funding of this study was provided by the Environmental Research and Technology Development Fund (4RF–1501 and 4–1304) of the Ministry of the Environment, Japan; a Grant-in-Aid for Young Scientists (A) (17H04996); a Grant-in-Aid for Research Fellows (19J21342) by The Japan Society for the Promotion of Science (JSPS); and the Marine Science R&D Programme (MSRDP-P03) supported by the National Research Foundation, Prime Minister’s Office, Singapore. ZR was supported by a JSPS short-term visiting Fellowship to the University of Miyazaki and samples from Western Australia were collected with the support of Woodside Energy on the Woodside Kimberley Collection Project.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Eriko Nagahiro for her assistance for genetic analysis. DM would like to thank Marco Casartelli and Julian Sitemba for their help in laboratory activities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.714662/full#supplementary-material
Supplementary Figure 1 | A bar plot of Heliopora sp. estimated by STRUCTURE ver. 2.3.4 assuming the number of cluster K = 3 using 225 SNPs obtained from the Stacks (r = 0.9 with the option –write_single_snp). Ten independent runs based on 200,000 burn-in followed by 200,000 Markov chain Monte Carlo (MCMC) replications were conducted using admixture models assuming correlated allele frequencies among 84 samples with uniform prior. The x-axis indicates each individual for Heliopora sp. that were classified into three subclades (sp.1–sp.3) in the phylogenetic tree (Figure 1). Different colors of the y-axis indicate the probability of assignment to different clusters. Red allows indicate possible hybrid individuals.
Supplementary Table 1 | Summary of sample information in this study: name of sampling country “Country,” name of sampling region “Region,” name of sampling site “Location,” longitude and latitude “Coordinates,” number of collected samples “Ns” and analyzed samples “Na,” ratio of assigned possible clones within each population “Rc,” name of Heliopora species group “Spp. group” and subclade “Subclade,” observed growth forms within each subclade “Major form (minor form),” and sampling years with published references “Year (References).”
References
Abe, M., Ohno, M., Kurozumi, T., Goto, T., Suzuki, R., Hasegawa, H., et al. (2008). Report of the Survey of Heliopora coerulea Communities in Oura Bay, Okinawa (in Japanese).
Altieri, A. H., Harrison, S. B., Seemann, J., Collin, R., Diaz, R. J., and Knowlton, N. (2017). Tropical dead zones and mass mortalities on coral reefs. Proc. Natl. Acad. Sci. U.S.A. 114, 3660–3665. doi: 10.1073/pnas.1621517114
Arrigoni, R., Berumen, M. L., Mariappan, K. G., Beck, P. S. A., Hulver, A. M., Montano, S., et al. (2020). Towards a rigorous species delimitation framework for scleractinian corals based on RAD sequencing: the case study of Leptastrea from the Indo-Pacific. Coral Reefs 39, 1001–1025. doi: 10.1007/s00338-020-01924-8
Babcock, R. (1990). Reproduction and development of the blue coral Heliopora coerulea (Alcyonaria: Coenothecalia). Mar. Biol. 104, 475–481. doi: 10.1007/bf01314352
Babcock, R. C. (1992). Workshop on Coral and Fish Recruitment. Manila, PH: University of the Philippines.
Babcock, R. C., Baird, A. H., Piromvaragorn, S., Thomson, D. P., and Willis, B. L. (2003). Identification of scleractinian coral recruits from Indo-Pacific reefs. Zool. Stud. 42, 211–226.
Baird, N. A., Etter, P. D., Atwood, T. S., Currey, M. C., Shiver, A. L., Lewis, Z. A., et al. (2008). Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3:e3376. doi: 10.1371/journal.pone.0003376
Bay, L., Choat, J. H., Herwerden, L., and Robertson, D. R. (2004). High genetic diversities and complex genetic structure in an Indo Pacific tropical reef fish (Chlorurus sordidus): evidence of an unstable evolutionary past? Mar. Biol. 144, 757–767. doi: 10.1007/s00227-003-1224-3
Benzie, J. A. H. (1999). Genetic structure of coral reef organisms: ghosts of dispersal past. Am. Zool. 39, 131–145. doi: 10.1093/icb/39.1.131
Benzie, J. A. H., Ballment, E., Forbes, A. T., Demetriades, N. T., Sugama, K., and Haryanti, M. S. (2002). Mitochondrial DNA variation in Indo-Pacific populations of the giant tiger prawn, Penaeus monodon. Mol. Ecol. 11, 2553–2569. doi: 10.1046/j.1365-294x.2002.01638.x
Bickford, D. P., Lohman, D. J., Sodhi, N. S., Ng, P. K. L., Meier, R., Winker, K., et al. (2007). Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22, 148–155. doi: 10.1016/j.tree.2006.11.004
Bilewitch, J. P., and Degnan, S. M. (2011). A unique horizontal gene transfer event has provided the octocoral mitochondrial genome with an active mismatch repair gene that has potential for an unusual self-contained function. BMC Evol. Biol. 11:228. doi: 10.1186/1471-2148-11-228
Birkeland, C., and Lucas, J. S. (1990). Acanthaster planci: Major Management Problem of Coral Reefs. West Palm Beach: CRC Press.
Bouillon, J., and Houvenaghel-Crévecoeur, N. (1970). Etude monographique du genre Heliopora de Blainville (Cenothecalia - Alcyonaria - Coelenterata). Ann. Mus. Roy. Afr. Centr. Sci. Zool. 178, 1–83.
Bowen, B. W., Rocha, L. A., Toonen, R. J., and Karl, S. A. (2013). The origins of tropical marine biodiversity. Trends Ecol. Evol. 28, 359–366. doi: 10.1016/j.tree.2013.01.018
Brady, A. K., Hilton, J. D., and Vize, P. D. (2009). Coral spawn timing is a direct response to solar light cycles and is not an entrained circadian response. Coral Reefs 28, 677–680. doi: 10.1007/s00338-009-0498-4
Bruno, J. F., Petes, L. E., Harvell, C. D., and Hettinger, A. (2003). Nutrient enrichment can increase the severity of coral diseases. Ecol. Lett. 6, 1056–1061. doi: 10.1046/j.1461-0248.2003.00544.x
Bruno, J. F., Selig, E. R., Casey, K. S., Page, C. A., Willis, B. L., Harvell, C. D., et al. (2007). Thermal stress and coral cover as drivers of coral disease outbreaks. PLoS Biol. 5:e124. doi: 10.1371/journal.pbio.0050124
Burge, C. A., Mark Eakin, C., Friedman, C. S., Froelich, B., Hershberger, P. K., Hofmann, E. E., et al. (2014). Climate change influences on marine infectious diseases: implications for management and society. Ann. Rev. Mar. Sci. 6, 249–277. doi: 10.1146/annurev-marine-010213-135029
Bushnell, B. (2017). BBTools Software Package. Available online at: http://sourceforge.net/projects/bbmap (accessed November 13, 2019).
Carpenter, K. E., Abrar, M., Aeby, G., Aronson, R. B., Banks, S., Bruckner, A., et al. (2008). One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science 321, 560–563.
Carpenter, K. E., Barber, P. H., Crandall, E. D., Ablan-Lagman, M. C. A., Ambariyanto, M., Mahardika, G. N., et al. (2011). Comparative phylogeography of the coral triangle and implications for marine management. J. Mar. Biol. 2011, 1–14. doi: 10.1155/2011/396982
Catchen, J. M., Amores, A., Hohenlohe, P., Cresko, W., and Postlethwait, J. H. (2011). Stacks: building and genotyping loci de novo from short-read sequences. G3 Genes Genom. Genet. 1, 171–182. doi: 10.1534/g3.111.000240
Cesar, H., Burke, L., and Pet-Soede, L. (2003). The Economics of Worldwide Coral Reef Degradation. Arnhem: Cesar Environmental Economics Consulting.
Colgan, M. W. (1984). “The cretaceous coral Heliopora (Octocorallia, Coenothecalia)-a common indo-pacific reef builder,” in Living Fossils, eds N. Eldredge and S. M. Stanley (Berlin: Springer).
Crowder, C. M., Meyer, E., Fan, T. Y., and Weis, V. M. (2017). Impacts of temperature and lunar day on gene expression profiles during a monthly reproductive cycle in the brooding coral Pocillopora damicornis. Mol. Ecol. 26, 3913–3925. doi: 10.1111/mec.14162
Dana, J. D. (1846). Zoophytes. United States Exploring Expedition V.7. Washington: Smithsonian libraries, 740.
Darling, E. S., Alvarez-Filip, L., Oliver, T. A., McClanahan, T. R., and Côté, I. M. (2012). Evaluating life-history strategies of reef corals from species traits. Ecol. Lett. 15, 1378–1386. doi: 10.1111/j.1461-0248.2012.01861.x
de Oliveira Soares, M., Matos, E., Lucas, C., Rizzo, L., Allcock, L., and Rossi, S. (2020). Microplastics in corals: an emergent threat. Mar. Pollut. Bull. 161:111810. doi: 10.1016/j.marpolbul.2020.111810
Eguchi, M. (1948). Fossil Helioporidae from Japan and the South Sea Islands. J. Paleont. 22, 362–364.
Erftemeijer, P. L. A., Riegl, B., Hoeksema, B. W., and Todd, P. A. (2012). Environmental impacts of dredging and other sediment disturbances on corals: a review. Mar. Pollut. Bull. 64, 1737–1765. doi: 10.1016/j.marpolbul.2012.05.008
Eyre, B. D., Cyronak, T., Drupp, P., De Carlo, E. H., Sachs, J. P., and Andersson, A. J. (2018). Coral reefs will transition to net dissolving before end of century. Science 359, 908–911. doi: 10.1126/science.aao1118
Fabricius, K., and Alderslade, P. (2001). Soft Corals and Sea Fans: a Comprehensive Guide to the Tropical Shallow-Water Genera of the Central-West Pacific, the Indian Ocean and the Red Sea. Townsville, AU: Australian Institute of Marine Science.
Fisher, R., O’Leary, R. A., Low-Choy, S., Mengersen, K., Knowlton, N., Brainard, R. E., et al. (2015). Species richness on coral reefs and the pursuit of convergent global estimates. Curr. Biol. 25, 500–505. doi: 10.1016/j.cub.2014.12.022
Flot, J. F., Magalon, H., Cruaud, C., Couloux, A., and Tillier, S. (2008). Patterns of genetic structure among Hawaiian corals of the genus Pocillopora yield clusters of individuals that are compatible with morphology. C. R. Biol. 331, 239–247. doi: 10.1016/j.crvi.2007.12.003
Forsman, Z. H., Barshis, D. J., Hunter, C. L., and Toonen, R. J. (2009). Shape-shifting corals: molecular markers show morphology is evolutionarily plastic in Porites. BMC Evol. Biol. 9:45. doi: 10.1186/1471-2148-9-45
Forsman, Z. H., Knapp, I. S. S., Tisthammer, K., Eaton, D. A. R., Belcaid, M., and Toonen, R. J. (2017). Coral hybridization or phenotypic variation? Genomic data reveal gene flow between Porites lobata and P. compressa. Mol. Phylogenet. Evol. 111, 132–148. doi: 10.1016/j.ympev.2017.03.023
Fukami, H., Omori, M., Shimoike, K., Hayashibara, T., and Hatta, M. (2003). Ecological and genetic aspects of reproductive isolation by different spawning times in Acropora corals. Mar. Biol. 142, 679–684. doi: 10.1007/s00227-002-1001-8
Gaither, M. R., Bowen, B. W., Bordenave, T. R., Rocha, L. A., Newman, S. J., Gomez, J. A., et al. (2011). Phylogeography of the reef fish Cephalopholis argus (Epinephelidae) indicates Pleistocene isolation across the Indo-Pacific Barrier with contemporary overlap in the Coral Triangle. BMC Evol. Biol. 11:189. doi: 10.1186/1471-2148-11-189
Gordon, A., and Hannon, G. J. (2012). FASTX-Toolkit. FASTQ/A Short-Reads Pre-Processing Tools. Available online at: http://hannonlab.cshl.edu/fastx_toolkit/ (accessed November 13, 2019).
Hall, R., and Holloway, J. D. (1998). Biogeography and Geological Evolution of SE Asia. Palo Alto, CA: Backhuys.
Halpern, B. S., Walbridge, S., Selkoe, K. A., Kappel, C. V., Micheli, F., D’Agrosa, C., et al. (2008). A global map of human impact on marine ecosystems. Science 319, 948–952.
Harii, S., and Kayanne, H. (2003). Larval dispersal, recruitment, and adult distribution of the brooding stony octocoral Heliopora coerulea on Ishigaki Island, southwest Japan. Coral Reefs 22, 188–196. doi: 10.1007/s00338-003-0302-9
Harii, S., Kayanne, H., Takigawa, H., Hayashibara, T., and Yamamoto, M. (2002). Larval survivorship, competency periods and settlement of two brooding corals, Heliopora coerulea and Pocillopora damicornis. Mar. Biol. 141, 39–46. doi: 10.1007/s00227-002-0812-y
Herrera, S., and Shank, T. M. (2016). RAD sequencing enables unprecedented phylogenetic resolution and objective species delimitation in recalcitrant divergent taxa. Mol. Phylogenet. Evol. 100, 70–79. doi: 10.1016/j.ympev.2016.03.010
Hirai, J. (2019). Insights into reproductive isolation within the pelagic copepod Pleuromamma abdominalis with high genetic diversity using genome-wide SNP data. Mar. Biol. 167:1. doi: 10.1007/s00227-019-3618-x
Hoegh-Guldberg, O. (2011). Coral reef ecosystems and anthropogenic climate change. Reg. Environ. Chang. 11, S215–S227.
Hoegh-Guldberg, O., and Bruno, J. F. (2010). The impact of climate change on the world’s marine ecosystems. Science 328, 1523–1528. doi: 10.1126/science.1189930
Hoegh-Guldberg, O., Mumby, P. J., Hooten, A. J., Steneck, R. S., Greenfield, P., Gomez, E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742. doi: 10.1126/science.1152509
Huang, D., Benzoni, F., Arrigoni, R., Baird, A. H., Berumen, M. L., Bouwmeester, J., et al. (2014). Towards a phylogenetic classification of reef corals: the Indo-Pacific genera Merulina, Goniastrea and Scapophyllia (Scleractinia, Merulinidae). Zool. Scripta 43, 531–548. doi: 10.1111/zsc.12061
Huang, W., Chen, M., Song, B., Deng, J., Shen, M., Chen, Q., et al. (2020). Microplastics in the coral reefs and their potential impacts on corals: a mini-review. Sci. Total Environ. 762:143112. doi: 10.1016/j.scitotenv.2020.143112
Hughes, T. P., Anderson, K. D., Connolly, S. R., Heron, S. F., Kerry, J. T., Lough, J. M., et al. (2018). Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359, 80–83. doi: 10.1126/science.aan8048
Hughes, T. P., Baird, A. H., Bellwood, D. R., Card, M., Connolly, S. R., Folke, C., et al. (2003). Climate change, human impacts, and the resilience of coral reefs. Science 301, 929–933. doi: 10.1126/science.1085046
Hui, M., Kraemer, W. E., Seidel, C., Nuryanto, A., Joshi, A., and Kochzius, M. (2016). Comparative genetic population structure of three endangered giant clams (Cardiidae: Tridacna species) throughout the Indo-West Pacific: implications for divergence, connectivity and conservation. J. Mollusc. Stud. 82, 403–414. doi: 10.1093/mollus/eyw001
Iguchi, A., Yoshioka, Y., Forsman, Z. H., Knapp, I. S., Toonen, R. J., Hongo, Y., et al. (2019). RADseq population genomics confirms divergence across closely related species in blue coral (Heliopora coerulea). BMC Evol. Biol. 19:187. doi: 10.1186/s12862-019-1522-0
Isomura, N., Yamauchi, C., Takeuchi, Y., and Takemura, A. (2013). Does dopamine block the spawning of the acroporid coral Acropora tenuis? Sci. Rep. 3:2649.
Johnston, E. C., Forsman, Z. H., Flot, J. F., Schmidt-Roach, S., Pinzón, J. H., Knapp, I. S. S., et al. (2017). A genomic glance through the fog of plasticity and diversification in Pocillopora. Sci. Rep. 7:5991. doi: 10.1038/s41598-017-06085-3
Jombart, T. (2008). adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. doi: 10.1093/bioinformatics/btn129
Jombart, T., and Ahmed, I. (2011). adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071. doi: 10.1093/bioinformatics/btr521
Jombart, T., Devillard, S., and Balloux, F. (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11:94. doi: 10.1186/1471-2156-11-94
Keith, S. A., Baird, A. H., Hughes, T. P., Madin, J. S., and Connolly, S. R. (2013). Faunal breaks and species composition of Indo-Pacific corals: the role of plate tectonics, environment and habitat distribution. Proc. R. Soc. Lond. 280:20130818. doi: 10.1098/rspb.2013.0818
Keyse, J., Crandall, E. D., Toonen, R. J., and Treml, E. A. (2014). The scope of published population genetic data for Indo-Pacific marine fauna and future research opportunities in the region. Bull. Mar. Sci. 90, 47–78. doi: 10.5343/bms.2012.1107
Knowlton, N. (1993). Sibling species in the sea. Annu. Rev. Ecol. Syst. 24, 189–216. doi: 10.1146/annurev.es.24.110193.001201
Knowlton, N., Mate, J. L., Guzman, H. M., Rowan, R., and Jara, J. (1997). Direct evidence for reproductive isolation among the three species of the Montastraea annularis complex in Central America (Panama and Honduras). Mar. Biol. 127, 705–711. doi: 10.1007/s002270050061
Lessios, H. A., Kessing, B. D., and Pearse, J. S. (2001). Population structure and speciation in tropical seas: global phylogeography of the sea urchin Diadema. Evolution 55, 955–975. doi: 10.1554/0014-3820(2001)055[0955:psasit]2.0.co;2
Levitan, D. R., Fogarty, N. D., Jara, J., Lotterhos, K. E., and Knowlton, N. (2011). Genetic, spatial, and temporal components of precise spawning synchrony in reef building corals of the Montastraea annularis species complex. Evolution 65, 1254–1270. doi: 10.1111/j.1558-5646.2011.01235.x
Levy, O., Appelbaum, L., Leggat, W., Gothlif, Y., Hayward, D. C., Miller, D. J., et al. (2007). Light-responsive cryptochromes from a simple multicellular animal, the coral Acropora millepora. Science 318, 467–470. doi: 10.1126/science.1145432
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows - Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lischer, H. E. L., and Excoffier, L. (2012). PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 28, 298–299. doi: 10.1093/bioinformatics/btr642
Luzon, K. S., Lin, M. F., Lagman, M. C. A. A., Licuanan, W. R. Y., and Chen, C. A. (2017). Resurrecting a subgenus to genus: molecular phylogeny of Euphyllia and Fimbriaphyllia (order Scleractinia; family Euphylliidae; clade V). PeerJ 5:e4074. doi: 10.7717/peerj.4074
Marquez, L. M., van Oppen, M. J. H., Willis, B. L., and Yan, M. (2002). Sympatric populations of the highly cross-fertile coral species, Acropora hyacinthus and Acropora cytherea, are genetically distinct. Proc. R. Soc. B 269, 1289–1294. doi: 10.1098/rspb.2002.2014
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Marti-Puig, P., Forsman, Z. H., Haverkort-Yeh, R. D., Knapp, I. S., Maragos, J. E., and Toonen, R. J. (2014). Extreme phenotypic polymorphism in the coral genus Pocillopora: micro-morphology corresponds to mitochondrial groups, while colony morphology does not. Bull. Mar. Sci. 90, 211–231. doi: 10.5343/bms.2012.1080
McFadden, C. S., Haverkort-Yeh, R., Reynolds, A. M., Halász, A., Quattrini, A. M., Forsman, Z. H., et al. (2017). Species boundaries in the absence of morphological, ecological or geographical differentiation in the Red Sea Octocoral genus Ovabunda (Alcyonacea: Xeniidae). Mol. Phylogenet. Evol. 112, 174–184. doi: 10.1016/j.ympev.2017.04.025
Meeker, N. D., Hutchinson, S. A., Ho, L., and Trede, N. S. (2007). Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques 43, 610–614. doi: 10.2144/000112619
Meirmans, P. G. (2020). GENODIVE version 3.0: easy-to-use software for the analysis of genetic data of diploids and polyploids. Mol. Ecol. Resour. 20, 1126–1131. doi: 10.1111/1755-0998.13145
Miller, M. R., Dunham, J. P., Amores, A., Cresko, W. A., and Johnson, E. A. (2007). Rapid and cost- effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 17, 240–248. doi: 10.1101/gr.5681207
Minh, B. Q., Nguyen, M. A., and von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Moberg, F., and Folke, C. (1999). Ecological goods and services of coral reef ecosystems. Ecol. Econ. 29, 215–233. doi: 10.1016/s0921-8009(99)00009-9
Montalbetti, E., Saponari, L., Montano, S., Maggioni, D., Dehnert, I., Galli, P., et al. (2019). New insights into the ecology and corallivory of Culcita sp. (Echinodermata: Asteroidea) in the Republic of Maldives. Hydrobiologia 827, 353–365. doi: 10.1007/s10750-018-3786-6
Nakabayashi, A., Matsumoto, T., Kitano, Y. F., Nagai, S., and Yasuda, N. (2017). Discovery of the northernmost habitat of the blue coral Heliopora coerulea: possible range expansion due to climate change? Galaxea J. Coral Reef Stud. 18, 1–2. doi: 10.3755/galaxea.19.1_1
Nakajima, Y., Nishikawa, A., Iguchi, A., Nagata, T., Uyeno, D., Sakai, K., et al. (2017). Elucidating the multiple genetic lineages and population genetic structure of the brooding coral Seriatopora (Scleractinia: Pocilloporidae) in the Ryukyu Archipelago. Coral Reefs 36, 415–426. doi: 10.1007/s00338-017-1557-x
Nakajima, Y., Nishikawa, A., Iguchi, A., and Sakai, K. (2012). The population genetic approach delineates the species boundary of reproductively isolated corymbose acroporid corals. Mol. Phylogenet. Evol. 63, 527–531. doi: 10.1016/j.ympev.2012.01.006
Obura, D. O. (2012). The diversity and biogeography of Western Indian Ocean reef-building corals. PLoS One 7:e45013. doi: 10.1371/journal.pone.0045013
Obura, D. O. (2016). An Indian Ocean centre of origin revisited: palaeogene and neogene influences defining a biogeographic realm. J. Biogeogr. 43, 229–242. doi: 10.1111/jbi.12656
Ohki, S., Kowalski, R. K., Kitanobo, S., and Morita, M. (2015). Changes in spawning time led to the speciation of the broadcast spawning corals Acropora digitifera and the cryptic species Acropora sp. 1 with similar gamete recognition systems. Coral Reefs 34, 1189–1198. doi: 10.1007/s00338-015-1337-4
Oldach, M. J., Workentine, M., Matz, M. V., Fan, T. Y., and Vize, P. D. (2017). Transcriptome dynamics over a lunar month in a broadcast spawning acroporid coral. Mol. Ecol. 26, 2514–2526. doi: 10.1111/mec.14043
Omori, M. (2011). Degradation and restoration of coral reefs: experience in Okinawa. Jpn. Mar. Biol. Res. 7, 3–12. doi: 10.1080/17451001003642317
Pallas, P. S. (1766). Elenchus Zoophytorum Sistens Generum Adumbrationes Generaliores et Speciarum Cognitarum Succintas Descriptiones cum Selectis Auctorus Synonymis. Hagae-Comitum: Petrum van Cleef, 1–451.
Palumbi, S. R. (1994). Genetic divergence, reproductive isolation, and marine speciation. Annu. Rev. Ecol. Syst. 25, 547–572. doi: 10.1146/annurev.es.25.110194.002555
Palumbi, S. R., Grabowsky, G., Duda, T., Geyer, L., and Tachino, N. (1997). Speciation and population genetic structure in tropical Pacific sea urchins. Evolution 51, 1506–1517. doi: 10.1111/j.1558-5646.1997.tb01474.x
Pante, E., Abdelkrim, J., Viricel, A., Gey, D., France, S. C., Boisselier, M. C., et al. (2015). Use of RAD sequencing for delimiting species. Heredity 114, 450–459. doi: 10.1038/hdy.2014.105
Park, J. S., Takayama, K., Suyama, Y., and Choi, B. H. (2019). Distinct phylogeographic structure of the halophyte Suaeda malacosperma (Chenopodiaceae/Amaranthaceae), endemic to Korea-Japan region, influenced by historical range shift dynamics. Plant Syst. Evol. 305, 193–203. doi: 10.1007/s00606-018-1562-8
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Pillans, B., Chappell, J., and Naish, T. R. (1998). A review of the Milankovitch climatic beat: template for Plio-Peistocene sea-level changes and sequence stratigraphy. Sediment. Geol. 122, 5–21. doi: 10.1016/S0037-0738(98)00095-5
Pinzón, J. H., Sampayo, E., Cox, E., Chauka, L. J., Chen, C. A., Voolstra, C. R., et al. (2013). Blind to morphology: genetics identifies several widespread ecologically common species and few endemics among Indo-Pacific cauliflower corals (Pocillopora, Scleractinia). J. Biogeogr. 40, 1595–1608. doi: 10.1111/jbi.12110
Planck, R. J., McAllister, D. E., and McAllister, A. T. (1988). Shiraho Coral Reef and The Proposed New Ishigaki Island Airport, Japan. Gland: IUCN.
Quattrini, A. M., Rodriguez, E., Faircloth, B. C., Cowman, P. F., Brugler, M. R., Farfan, G. A., et al. (2020). Paleoclimate ocean conditions shaped the evolution of corals and their skeletons through deep time. Nat. Ecol. Evol. 4, 1531–1538. doi: 10.1038/s41559-020-01291-1
Quattrini, A. M., Wu, T., Soong, K., Jeng, M. S., Benayahu, Y., and McFadden, C. S. (2019). A next generation approach to species delimitation reveals the role of hybridization in a cryptic species complex of corals. BMC Evol. Biol. 19:116. doi: 10.1186/s12862-019-1427-y
Rambaut, A. (2012). FigTree: Tree Figure Drawing Tool Version 1.4.4. Available online at: http://tree.bio.ed.ac.uk/software/figtree (accessed November 13, 2019).
Richards, Z. T., Berry, O., and van Oppen, M. J. (2016). Cryptic genetic divergence within threatened species of Acropora coral from the Indian and Pacific Oceans. Conserv. Genet. 17, 577–591. doi: 10.1007/s10592-015-0807-0
Richards, Z. T., Haines, L., Scaps, P., and Ader, D. (2020). New records of Heliopora hiberniana from SE Asia and the Central Indian Ocean. Diversity 12:328. doi: 10.3390/d12090328
Richards, Z. T., Miller, D. J., and Wallace, C. C. (2013). Molecular phylogenetics of geographically restricted Acropora species: implications for threatened species conservation. Mol. Phylogenet. Evol. 69, 837–851. doi: 10.1016/j.ympev.2013.06.020
Richards, Z. T., Yasuda, N., Kikuchi, T., Foster, T., Mitsuyuki, C., Stat, M., et al. (2018). Integrated evidence reveals a new species in the ancient blue coral genus Heliopora (Octocorallia). Sci. Rep. 8:15875.
Ridgway, T., and Gates, D. (2006). Why are there so few genetic markers available for coral population analyses? Symbiosis 41, 1–7. doi: 10.1093/oso/9780190907976.003.0001
Rochette, N. C., Rivera-Colon, A. G., and Catchen, J. M. (2019). Stacks 2: analytical methods for paired end sequencing improve RADseq-based population genomics. Mol. Ecol. 28, 4737–4754. doi: 10.1111/mec.15253
Rosser, N. L. (2015). Asynchronous spawning in sympatric populations of a hard coral reveals cryptic species and ancient genetic lineages. Mol. Ecol. 24, 5006–5019. doi: 10.1111/mec.13372
Rosser, N. L., Thomas, L., Stankowski, S., Richards, Z. T., Kennington, W. J., and Johnson, M. S. (2017). Phylogenomics provides new insight into evolutionary relationships and genealogical discordance in the reef-building coral genus Acropora. Proc. R. Soc. B Biol. Sci. 284:20162182. doi: 10.1098/rspb.2016.2182
Rowe, H. C., Renaut, S., and Guggisberg, A. (2011). RAD in the realm of next-generation sequencing technologies. Mol. Ecol. 20, 3499–3502.
Saito, Y., Ueno, M., Kiatano, Y. F., and Yasuda, N. (2015). Potential of different reproductive timing between sympatric Heliopora coerulea lineages southeast of Iriomote Island, Japan. Bul. Mar. Sci. 91, 397–398. doi: 10.5343/bms.2015.1024
Schettino, A., and Turco, E. (2011). Tectonic history of the western Tethys since the Late Triassic. Geol. Soc. Am. Bull. 123, 89–105. doi: 10.1130/b30064.1
Schmidt-Roach, S., Lundgren, P., Miller, K. J., Gerlach, G., Noreen, A. M. E., and Andreakis, N. (2013). Assessing hidden species diversity in the coral Pocillopora damicornis from Eastern Australia. Coral Reefs 32, 161–172. doi: 10.1007/s00338-012-0959-z
Schott, F. A., and McCreary, J. P. (2001). The monsoon circulation of the Indian Ocean. Prog. Oceanogr. 51, 11–23.
Schott, F. A., Xie, S. P., and McCreary, J. P. Jr. (2009). Indian Ocean circulation and climate variability. Rev. Geophys. 47:RG1002. doi: 10.1029/2007RG000245
Shafer, A. B. A., Peart, C. R., Tusso, S., Maayan, I., Brelsford, A., Wheat, C. W., et al. (2017). Bioinformatic processing of RAD-seq data dramatically impacts downstream population genetic inference. Methods Ecol. Evol. 8, 907–917. doi: 10.1111/2041-210x.12700
Shearer, T. L., van Oppen, M. J. H., Romano, S. L., and Worheide, G. (2002). Slow mitochondrial DNA sequence evolution in the Anthozoa (Cnidaria). Mol. Ecol. 11, 2475–2487. doi: 10.1046/j.1365-294x.2002.01652.x
Souter, P. (2010). Hidden genetic diversity in a key model species of coral. Mar. Biol. 157, 875–885. doi: 10.1007/s00227-009-1370-3
Spalding, M. D., Ravilious, C., and Green, E. P. (2001). World Atlas of Coral Reefs. Prepared at the UNEP World Conservation Monitoring Centre. Berkeley, CA: University of California Press.
Suyama, Y., and Matsuki, Y. (2015). MIG-seq: an effective PCR-based method for genome-wide single-nucleotide polymorphism genotyping using the next-generation sequencing platform. Sci. Rep. 5:16963. doi: 10.1038/srep16963
Swierts, T., and Vermeij, M. J. (2016). Competitive interactions between corals and turf algae depend on coral colony form. PeerJ 4:e1984.
Szmant, A. M., Weil, E., Miller, M., and Colon, D. E. (1997). Hybridization within the species complex of the scleractinian coral Montastraea annularis. Mar. Biol. 129, 561–572. doi: 10.1007/s002270050197
Takata, K., Taninaka, H., Nonaka, M., Iwase, F., Kikuchi, T., Suyama, Y., et al. (2019). Multiplexed ISSR genotyping by sequencing distinguishes two precious coral species (Anthozoa: Octocorallia: Coralliidae) that share a mitochondrial haplotype. PeerJ 7:e7769. doi: 10.7717/peerj.7769
Takino, T., Watanabe, A., Motooka, S., Nadaoka, K., Yasuda, N., and Taira, M. (2010). Discovery of a large population of Heliopora coerulea at akaishi reef, Ishigaki Island, southwest Japan. Galaxea J. Coral Reef Stud. 12, 85–86. doi: 10.3755/galaxea.12.85
Tamaki, I., Yoichi, W., Matsuki, Y., Suyama, Y., and Mizuno, M. (2017). Inconsistency between morphological traits and ancestry of individuals in the hybrid zone between two Rhododendron japonoheptamerum varieties revealed by a genotyping-by-sequencing approach. Tree Genet. Genom. 13:4.
Taninaka, H., Bernardo, L. P. C., Saito, Y., Nagai, S., Ueno, M., Kitano, Y. F., et al. (2019). Limited fine-scale larval dispersal of the threatened brooding corals Heliopora spp. as evidenced by population genetics and numerical simulation. Conserv. Genet. 20, 1449–1463. doi: 10.1007/s10592-019-01228-7
Taninaka, H., Harii, S., Kagawa, H., Ueno, M., Kitano, Y. F., Saito, Y., et al. (2018). Estimation of the reproductive timing of two genetically different lineages of the blue coral Heliopora coerulea (Pallas, 1766) around Sekisei Lagoon. J. Jpn. Coral Reef Soc. 20, 39–51. doi: 10.3755/jcrs.20.39
Todd, P. A. (2008). Morphological plasticity in scleractinian corals. Biol. Rev. 83, 315–337. doi: 10.1111/j.1469-185x.2008.00045.x
van Oppen, M. J. H., McDonald, B. J., Willis, B., and Miller, D. J. (2001). The evolutionary history of the coral genus Acropora (Scleractinia, Cnidaria) based on a mitochondrial and a nuclear marker: reticulation, incomplete lineage sorting, or morphological convergence? Mol. Biol. Evol. 18, 1315–1329. doi: 10.1093/oxfordjournals.molbev.a003916
Veron, J. E. (1995). Corals in Space and Time: The Biogeography and Evolution of the Scleractinia. Sydney: UNSW Press.
Veron, J. E. N. (2000). Corals of the World, Vol. 1–3. Townsville, AU: Australian Institute of Marine Science and CRR.
Veron, J. E. N., Stafford-Smith, M., De Vantier, L., and Turak, E. (2015). Overview of distribution patterns of zooxanthellate Scleractinia. Front. Mar. Sci. 1:81. doi: 10.3389/fmars.2014.00081
Villanueva, R. D. (2016). Cryptic speciation in the stony octocoral Heliopora coerulea: temporal reproductive isolation between two growth forms. Mar. Biodivers. 46, 503–507. doi: 10.1007/s12526-015-0376-y
Vogler, C., Benzie, J., Lessios, H., Barber, P., and Worheide, G. (2008). A threat to coral reefs multiplied? Four species of crown-of-thorns starfish. Biol. Lett. 4, 696–699. doi: 10.1098/rsbl.2008.0454
Vollmer, S. V., and Palumbi, S. R. (2004). Testing the utility of internally transcribed spacer sequences in coral phylogenetics. Mol. Ecol. 13, 2763–2772. doi: 10.1111/j.1365-294x.2004.02265.x
Warner, P. A., van Oppen, M. J., and Willis, B. L. (2015). Unexpected cryptic species diversity in the widespread coral Seriatopora hystrix masks spatial-genetic patterns of connectivity. Mol. Ecol. 24, 2993–3008. doi: 10.1111/mec.13225
Wells, J. W. (1954). Recent corals of the Marshall Islands. U.S. Geol. Surv. Prof. Pap. 260, 285–486.
Wepfer, P. H., Nakajima, Y., Sutthacheep, M., Radice, V. Z., Richards, Z., Ang, P., et al. (2020). Evolutionary biogeography of the reef-building coral genus Galaxea across the Indo-Pacific ocean. Mol. Phylogenet. Evol. 151:106905. doi: 10.1016/j.ympev.2020.106905
Wepfer, P. H., Nakajima, Y., Sutthacheep, M., Radice, V. Z., Richards, Z., Ang, P., et al. (2021). Inclusivity is key to progressing coral biodiversity research: reply to comment by Bonito et al. 2021. Mol. Phylogenet. Evol. 6:107135. doi: 10.1016/j.ympev.2021.107135
Wilkinson, C. (2008). Status of Coral Reefs of the World. Townsville: Global Coral Reef Monitoring Network and Reef and Rain forest Research Centre, 296.
Williams, S. T., and Benzie, J. A. H. (1998). Evidence of a biogeographic break between populations of a high dispersal starfish: congruent regions within the Indo-West Pacific defined by color morphs, mtDNA and allozyme data. Evolution 52, 87–99. doi: 10.2307/2410923
Wörheide, G., Epp, L. S., and Macis, L. (2008). Deep genetic divergences among Indo-Pacific populations of the coral reef sponge Leucetta chagosensis (Leucettidae): founder effects, vicariance, or both? BMC Evol. Biol. 8:24. doi: 10.1186/1471-2148-8-24
Yasuda, N. (2018). “Distribution expansion and historical population outbreak patterns of crown-of-thorns starfish, Acanthaster planci sensu lato,” in Coral Reef Studies of Japan. Coral Reefs of the World, Japan From 1912 to 2015, Vol. 13, eds A. Iguchi and C. Hongo (Singapore: Springer), 125–148. doi: 10.1007/978-981-10-6473-9_9
Yasuda, N., Takino, T., Kimura, M., Lian, C., Nagai, S., and Nadaoka, K. (2010). “Genetic structuring across the reef crest in the threatened blue coral Heliopora coerulea (Helioporidae, Octacorallia) in Shiraho reef, Southwest Japan,” in Advances in Genetics Research, ed. K. V. Urbano (New York, NY: Nova Science Publishers, Inc), 315–324.
Yasuda, N., Taquet, C., Nagai, S., Fortes, M., Fan, T. Y., Harii, S., et al. (2015). Genetic diversity, paraphyly & incomplete lineage sorting of mtDNA, ITS2 snd microsatellite flanking region in closely related Heliopora species (Octocorallia). Mol. Phylogenet. Evol. 93, 161–171.
Yasuda, N., Taquet, C., Nagai, S., Fortes, M., Fan, T. Y., Phongsuwan, N., et al. (2014). Genetic structure and cryptic speciation in the threatened reef-building coral Heliopora coerulea along Kuroshio Current. Bull. Mar. Sci. 90, 233–255. doi: 10.5343/bms.2012.1105
Keywords: MIG-seq, single nucleotide polymorphism, Helioporadae, octocoral, species delimitation, coral reef, evolutionary relationships, species diversity
Citation: Taninaka H, Maggioni D, Seveso D, Huang D, Townsend A, Richards ZT, Tang S-L, Wada N, Kikuchi T, Yuasa H, Kanai M, De Palmas S, Phongsuwan N and Yasuda N (2021) Phylogeography of Blue Corals (Genus Heliopora) Across the Indo-West Pacific. Front. Mar. Sci. 8:714662. doi: 10.3389/fmars.2021.714662
Received: 25 May 2021; Accepted: 23 June 2021;
Published: 13 August 2021.
Edited by:
Wei Jiang, Guangxi University, ChinaReviewed by:
Snaebjörn Pálsson, University of Iceland, IcelandNicole De Voogd, Naturalis Biodiversity Center, Netherlands
Copyright © 2021 Taninaka, Maggioni, Seveso, Huang, Townsend, Richards, Tang, Wada, Kikuchi, Yuasa, Kanai, De Palmas, Phongsuwan and Yasuda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nina Yasuda, nina27@cc.miyazaki-u.ac.jp
 Hiroki Taninaka
Hiroki Taninaka Davide Maggioni
Davide Maggioni Davide Seveso
Davide Seveso Danwei Huang
Danwei Huang Abram Townsend7
Abram Townsend7  Sen-Lin Tang
Sen-Lin Tang Naohisa Wada
Naohisa Wada Taisei Kikuchi
Taisei Kikuchi Stéphane De Palmas
Stéphane De Palmas Nina Yasuda
Nina Yasuda