Molecular Phylogenetics and Evolution ( IF 4.1 ) Pub Date : 2023-01-25 , DOI: 10.1016/j.ympev.2023.107715 Robert Literman 1 , Amanda M Windsor 1 , Henry L Bart 2 , Elizabeth Sage Hunter 1 , Jonathan R Deeds 1 , Sara M Handy 1
|
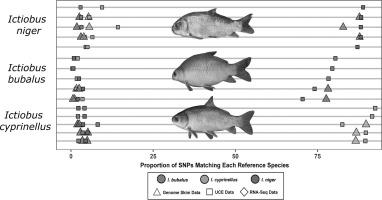
Consumption of buffalofish has been sporadically associated with Haff disease-like illnesses involving sudden onset muscle pain and weakness due to skeletal muscle rhabdomyolysis, but determination of precisely which species are associated with these illnesses has been impeded by a lack of species-specific DNA-based markers. Here, three closely related species of buffalofish native to the Mississippi River Basin (Ictiobus bubalus, Ictiobus cyprinellus and Ictiobus niger) that have previously proven genetically indistinguishable using both mitochondrial and nuclear single-locus sequencing were reliably discriminated using low-coverage whole genome sequencing (‘genome skimming’). Using 44 specimens representing the three species collected from the mid/upper (Missouri) and lower (Louisiana) regions of the species’ native ranges, the SISRS (Site Identification from Short Read Sequences) bioinformatics pipeline was adapted to (1) identify over 620Mbp of putatively homologous nuclear sequence data and (2) isolate over 140,000 single-nucleotide polymorphisms (SNPs) that supported accurate species delimitation, all without the use of a reference genome or annotation data. These sites were used to classify Ictiobus spp. samples with genome-skim data, along with a larger set (n = 67) where ultraconserved elements (UCEs) were sequenced. Analyses of whole mitochondrial data revealed more limited signal. Nearly all samples matched their purported species based on morphologic identification, but two Missouri samples morphologically identified as I. niger grouped with samples of I. bubalus, albeit with significant enrichment of I. niger SNPs. To our knowledge this is the first report of a DNA-based tool to reliably discriminate these three morphologically distinct species.
中文翻译:

使用低覆盖度全基因组测序(基因组略读)描绘三种渐渗的水牛鱼 (Ictiobus)
食用水牛鱼偶尔会引起类似哈夫病的疾病,包括由于骨骼肌横纹肌溶解症引起的突然发作的肌肉疼痛和虚弱,但由于缺乏基于物种特异性 DNA 的技术,无法准确确定哪些物种与这些疾病有关标记。在这里,原产于密西西比河流域的三种密切相关的水牛鱼种类(Ictiobus bubalus、Ictiobus cyprinellus和Ictiobus niger) 之前已证明使用线粒体和核单基因座测序在遗传上无法区分的细胞,使用低覆盖率全基因组测序(“基因组略读”)可靠地区分。使用代表从物种原生范围的中上/上部(密苏里州)和下部(路易斯安那州)地区收集的三个物种的 44 个标本,SISRS(短读取序列的站点识别)生物信息学管道适用于(1)识别超过 620Mbp推定的同源核序列数据,以及 (2) 分离超过 140,000 个支持准确物种定界的单核苷酸多态性 (SNP),所有这些都没有使用参考基因组或注释数据。这些站点用于对Ictiobus进行分类属 具有基因组脱脂数据的样本,以及对超保守元素 (UCE) 进行测序的更大集合 (n = 67)。对整个线粒体数据的分析揭示了更有限的信号。根据形态学鉴定,几乎所有样品都与其声称的物种相匹配,但两个密苏里州样品在形态学上被鉴定为I. niger与I. bubalus样品分组,尽管I. niger SNP 显着富集。据我们所知,这是基于 DNA 的工具的第一份报告,可以可靠地区分这三种形态不同的物种。



























 京公网安备 11010802027423号
京公网安备 11010802027423号